Figures & data
Figure 1 Templating specificity of the C. albicans prion strain conformation of the S17R Sup35NM chimera. The S17R Sup35NM chimera (Chim) and cysteine mutants thereof were assembled into prion amyloids and their templating specificity was analyzed using ThT fluorescence. The reported seeding activity of the chimeric Sup35NM fibrils is after subtraction of the contribution from the residual CaNM fibrils used to template the assembly of the Sup35NM chimera.
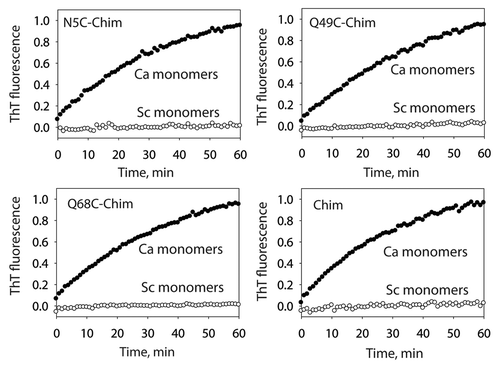
Figure 2 Solvent accessibility of cysteine residues in the C. albicans prion strain conformation of the Sup35NM chimera. (A) S17R Sup35NM chimera fibrils in the C. albicans strain conformation (as well as the corresponding monomers) were labeled using Lucifer yellow iodoacetamide. (B) Comparison of cysteine labeling of S17R and wild-type Sup35NM chimera fibrils in the S. cerevisiae domain. Each measurement is the average (±standard deviation) of at least three independent experiments.
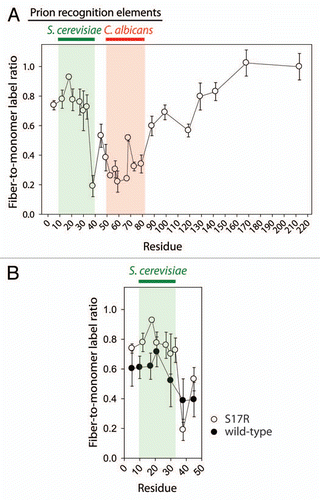
Figure 3 Impact of proline substitution mutants on the templating activity of the C. albicans strain conformation of the S17R Sup35NM chimera. S17R Sup35NM fibrils in the C. albicans strain conformation were evaluated for their activity to template proline substitution mutants of the Sup35NM chimera into prion fibrils. Each seeding rate (k) was calculated from the initial slope of the ThT measurements, and expressed as percent defect [100 ×(1 − kmut/kwt)], as described previously in reference Citation38.
![Figure 3 Impact of proline substitution mutants on the templating activity of the C. albicans strain conformation of the S17R Sup35NM chimera. S17R Sup35NM fibrils in the C. albicans strain conformation were evaluated for their activity to template proline substitution mutants of the Sup35NM chimera into prion fibrils. Each seeding rate (k) was calculated from the initial slope of the ThT measurements, and expressed as percent defect [100 ×(1 − kmut/kwt)], as described previously in reference Citation38.](/cms/asset/485a5ea8-f785-44a7-bdb5-f49ff44e2337/kprn_a_10916694_f0003.gif)
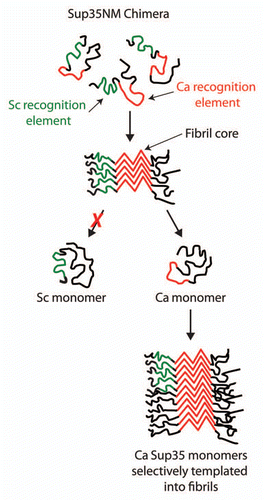
Figure 4 Selective folding of prion recognition elements within the Sup35NM chimera enciphers species-specific infectivity. The Sup35NM chimera contains two prion recognition sequences (green for the S. cerevisiae recognition element and red for C. albicans recognition element) that selectively nucleate each prion strain conformation. For the C. albicans strain conformation, we find that the Ca recognition sequence is folded within the prion amyloid core, while most of the Sc recognition sequence is excluded from the core. The resulting prion strain conformation is specific for infecting C. albicans Sup35 since its cognate recognition element is folded within the amyloid core and active for templating.