Figures & data
Figure 1 (A) Graphic representation of the behavioral changes in C57BL6 and albino Swiss mice models of prion disease. Left, middle and right columns correspond to burrowing, open field and rod bridge performances. Top and middle rows illustrate average values to C57BL6 and albino Swiss whereas bottom row illustrate two different subgroups (ME7-E) and (ME7-L) that start to behavioral deficits early and late respectively. (B) Cluster and discriminant analysis at 18 w.p.i. to demonstrate the presence of distinct sensitivity to ME7 agent. Note that on average C57BL6 start to change burrowing at 12 w.p.i. whereas albino Swiss mice only start 4 weeks later. ME7-E start at the same time as C57BL6, whereas ME7-L 10 weeks later. Open field activity starts to increase on average at 16 w.p.i. in C57Bl6 whereas in albino Swiss mice start 6 weeks later. C57BL6 and albino Swiss mice show the onset of impairments of motor coordination at the same time (18 w.p.i.). Despite statistical significant differences between ME7-animals and NBH-animals, cluster analysis did not detect subgroups in the open field and rod bridge performances.
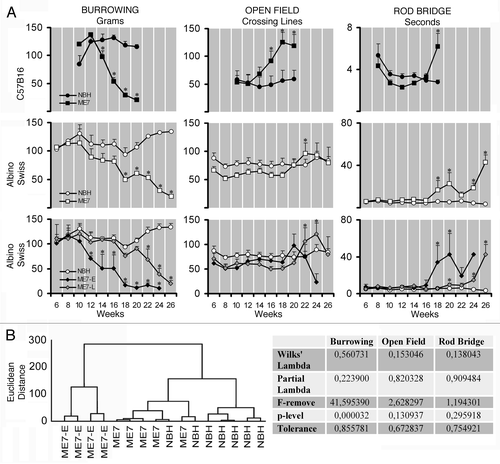
Figure 2 Photomicrographs of hippocampal region of ME7 subjects at 18 w.p.i. after Neu-N (A) and PrP (B) immunohistochemistry to illustrate vacuoles and PrPSc deposits (white arrows) respectively. Photomicrographs of ME7-animal (C) and NBH-animal (D) mossy fibers at 18 w.p.i. Black arrows indicate damaged axons with boutons tumefaction, and reduction of the axonal filling in the ME7 infected brain whereas in the NBH-animal these signs are not present. Scale bars: (A and B) 250 µm; (C and D) 25 µm.
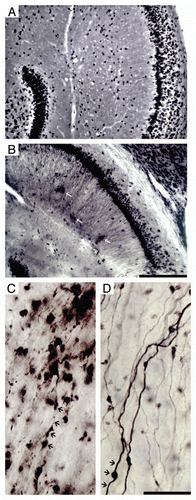
Figure 3 Photomicrographs of the polymorphic layer of dentate gyrus of ME7 subjects at 12 w.p.i. after Lycopersicum esculentum histochemistry (n = 9) to illustrate microglial activation early in the disease. Right column show early microglial activation in 4 infected subjects (ME7-E) as compared with 4 subjects without early microglia activation (left column, ME7-L). Scale bar: 25 µm.
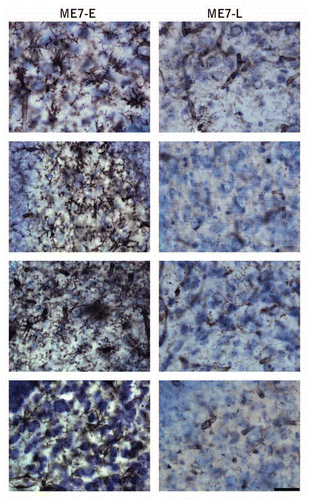
Figure 4 Top and bottom: low and high power photomicrographics of the polymorphic layer of dentate gyrus (PolDG) and CA1 of GFAP immunostained sections after 15 or 18 w.p.i. Graphic representations illustrate the mean values and error bars of the total number of astrocytes in the PolDG (left) and CA1 (middle) and of the activation index (right) of reactive astrocytes in CA1 and polymorphic layer. Activation index was estimated by the following equation AI18/NBH = (ME718w − NBH)/(ME718w + NBH) or AI18/15 = (ME718w − ME715w)/(ME718w + ME7 15w) or AI15/NBH = (ME715w − NBH)/(ME715w + NBH) where AI is the activation index in the period and ME718w, ME715w and NBH are the estimates of the number of objects of interest at 15 and 18 w.p.i. in each region for each experimental group. Significant differences are indicated by (*) and the probability values by p level; (*) = 0.05; (**) = 0.025; (***) = 0.01. Scale bars: Scale bars: low power 250 µm; high power 25 µm.
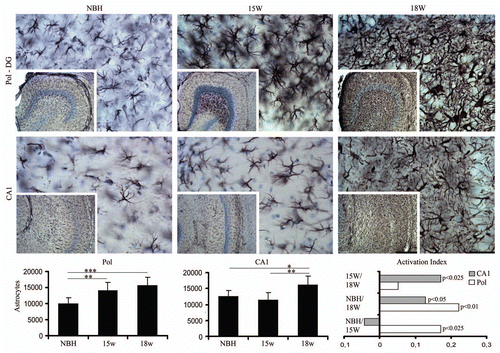
Figure 5 Top and bottom: low and high power photomicrographs of the polymorphic layer of dentate gyrus (Pol-DG) and CA1 of activated microglia stained with Lycopersicum esculentum histochemistry after 15 or 18 w.p.i. Graphic representations illustrate the mean values and error bars of the total number of activated microglia in the polymorphic layer of dentate gyrus (left) and CA1 (middle) and of the activation index (right) of microglia in CA1 and polymorphic layer. Activation index was estimated by the following equation AI18/NBH = (ME718w − NBH)/(ME718w + NBH) or AI18/15 = (ME718w − ME715w)/(ME718w + ME7 15w) or AI15/NBH = (ME715w − NBH)/(ME715w + NBH) where AI is the activation index in the period and ME718w, ME715w and NBH are the estimates of the number of objects of interest at 15 and 18 w.p.i. in each region for each experimental group. Significant differences are indicated by (*) and the probability values by p level; (*) = 0.05; (**) = 0.025; (***) = 0.01. Scale bars: Scale bars: low power 250 µm; high power 25 µm.
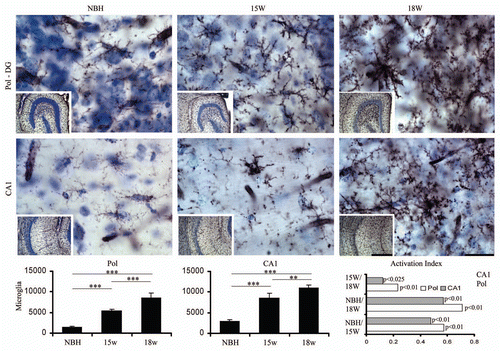
Figure 6 Septal region, low and high power, photomicrographs to illustrate GFAP (top) and Lycopersicum esculentum (bottom) reacted sections after 15 or 18 w.p.i. Graphic representations illustrate the mean values and error bars of the total number of astrocytes and activated microglia in the septal region and corresponding activation index. Activation index was estimated by the following equation AI18/NBH = (ME718w − NBH)/(ME718w + NBH) or AI18/15 = (ME718w − ME715w)/(ME718w + ME7 15w) or AI15/NBH = (ME715w − NBH)/(ME715w + NBH) where AI is the activation index in the period and ME718w, ME715w and NBH are the estimates of the number of objects of interest at 15 and 18 w.p.i. in each region for each experimental group. Significant differences are indicated by (*) and the probability values by p level; (*) = 0.05; (**) = 0.025; (***) = 0.01. Scale bars: Scale bars: low power 250 µm; high power 25 µm.
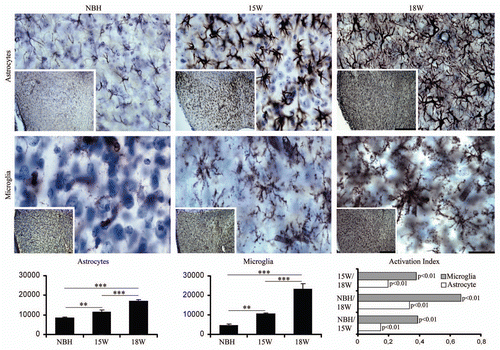
Figure 7 Top and middle: low and high power photomicrographs of CA1 and septum of sections stained for biotinylated Wisteria floribunda to demonstrate perineuronal nets after 15 or 18 w.p.i. of infected (15 w, 18 w) or normal brain (NBH) homogenates. Bottom: graphic representations of the mean and error bars estimations of the total number of perineuronal nets in CA1 (left) and septum (middle) and of the reduction index (right) of perineuronal nets in CA1 (white bars) and septum (grey bars). Reduction index was estimated by the following equation RINBH/18 = (ME718w − NBH)/(ME718w + NBH) or RI15/18 = (ME715w − ME718w)/(ME715w + ME7 18w) or RINBH/15 = (ME7NBH − ME715w)/(NBH + ME715w) where RI is the reduction index in the period and ME718w, ME715w and NBH are the estimations of the number of objects of interest at 15 and 18 w.p.i. of ME7 infected or normal brain (NBH) homogenates in each region for each experimental group. Significant differences are indicated by *p < 0.05 or **p < 0.01.
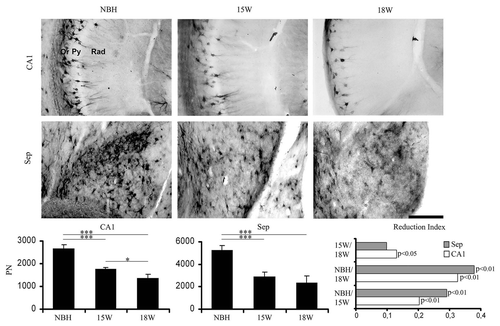
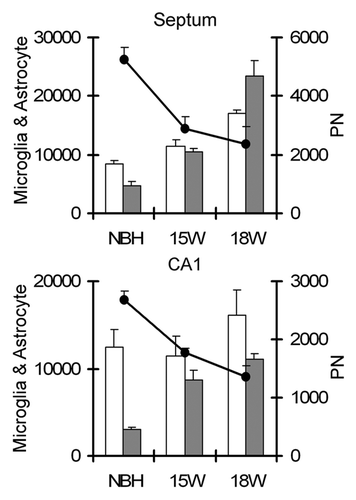
Figure 8 Overview of the total numbers of astrocytes, microglia and PN nets estimations in the septum (top) and CA1 (bottom). Note that the total number of perineuronal nets (PN) decrease along the disease progression whereas astrocytes and microglia increase. White and black columns correspond to astrocytes and microglia respectively whereas data points correspond to PN nets estimations.