Figures & data
Figure 1. Quantitative RT-PCR analysis of BDG1, SHN1, SHN2, and SHN3 expression in leaves. The mean of the CT (cycle threshold) values of the reference gene (eIF1α) was subtracted from the respective CT value of the gene of interest. Subsequently, differences were subtracted from the wild-type value. Numbers give fold changes in comparison to the wild type. Error: SE of gene of interest in three technical replicates. (A) Relative transcript levels in 19-d-old plants grown in half-concentrated MS medium. (B) Relative transcript levels in 4-week-old soil-grown plants.
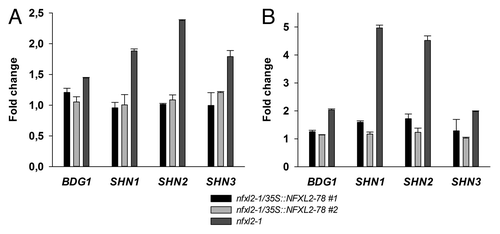
Figure 2. Putative X-box-like sequences in BDG1 and SHINE promoters. The X-box sequence in MHC class II promoters and the X-box-like sequence preceded by the overlapping E-box within the hTERT promoter are shown in comparison to sequences identified in the BDG1, SHN1, SHN2, and SHN3 promoters. A consensus sequence matching the depicted Arabidopsis sequences is given at the bottom. Highly conserved residues include the ACGT sequence. This sequence is part of the human E-box and resembles the core sequence of the ABA response element (ABRE).Citation20 The conserved AA and TG nucleotides were among the five residues that were shown to cause diminished NFX1–91 binding to the hTERT promoter when being mutated.Citation15 X: any nucleotide. S: C or G.
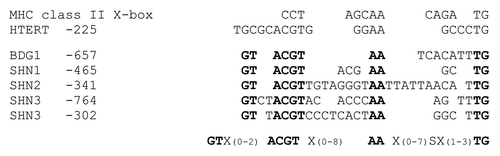
Figure 3. In vivo binding of GFP-NFXL2–78 to BDG1, SHN1, SHN2, and SHN3 promoters and analysis of GFP-NFXL2–78 transcript levels. (A) PCR amplification of promoter fragments after ChIP with an anti-GFP antibody in leaf extracts of 4-week-old soil-grown plants. Numbers give fold changes ± SE in three technical replicates (two nfxl2–1/35S::GFP-NFXL2–78 lines plus vs. minus anti-GFP antibody). (B) Semiquantitative RT-PCR was used to determine GFP-NFXL2–78 mRNA levels in 4-week-old soil-grown plants. As an internal control, the same cDNAs were used to quantify TPT transcript levels. The GFP-NFXL2–78 and TPT transcripts were amplified by PCR for 24/27 and 24 cycles, respectively. The PCR products were separated on a 1.5% agarose gel. 1, wild type; 2, nfxl2–1; 3, 35S::GFP-NFXL2–78 #1; 4, 35S::GFP-NFXL2–78 #2.
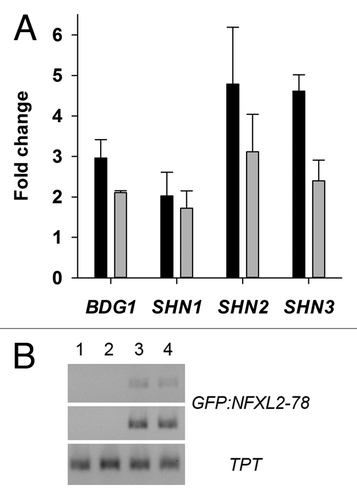
Figure 4. Analysis of cuticle permeability. (A) Chlorophyll leaching assays with mature rosette leaves. Rosette leaves of 4-week-old soil-grown plants were immersed in 80% ethanol for different time periods. Results are given as mean ± SE (B) Toluidine-blue staining of rosette leaves. Representative rosette leaves are shown after staining with an aqueous solution (0.05%) of toluidine blue. Left: Plants grown under aseptic conditions. Right: Plants grown in soil.
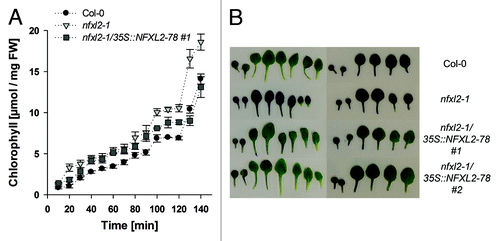
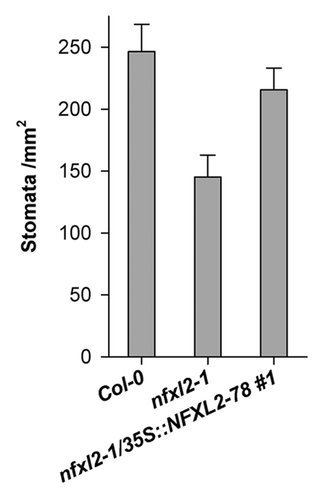
Figure 5. Stomatal density of mature abaxial leaf blades. Stomatal density was determined in mature rosette leaves of 4-week-old soil-grown plants. Results are given as mean ± SE. Stomatal density of nfxl2–1 plants is significantly different from the wild type (t test, p < 0.01).