Figures & data
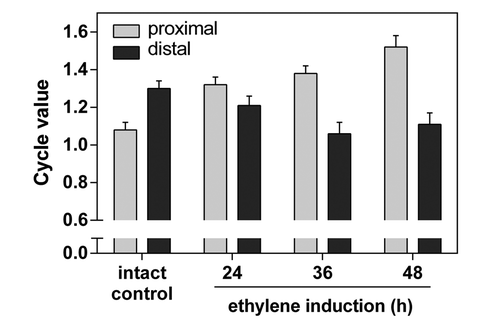
Figure 1. Model of the processes occurring in the AZ. A major process on the proximal side of the AZ is endoreduplication, presumably associated with extensive membrane trafficking. Its ultrastructure is characterized by electron-lucent ameboidal-shaped nuclei, a well developed Golgi apparatus and endoplasmic reticulum connected with an elaborate plasmodesmal system, pronounced membrane invaginations, and multivesicular and paramural bodies. In addition, polygalacturonases - enzymes involved in the execution of cell separationCitation4,Citation18 are also located at the proximal side of the AZ. At the distal side of the AZ programmed cell death occurs. It is identified by loss of cell viability, altered nuclear morphology, DNA fragmentation, elevated levels of reactive oxygen species, and elevated enzymatic activities and expression of PCD-associated genes, including genes encoding LX ribonuclease, nuclease TBN1, cysteine and serine proteases, tomato β-1,4-endoglucanase, which is involved in cell-wall metabolism; and 1-aminocyclopropane-1-carboxylic acid synthase, involved in ethylene biosynthesis.Citation5 Arrow-ended and blunt-ended lines represent process induction and repression, respectively, dashed lines represent putative relations.
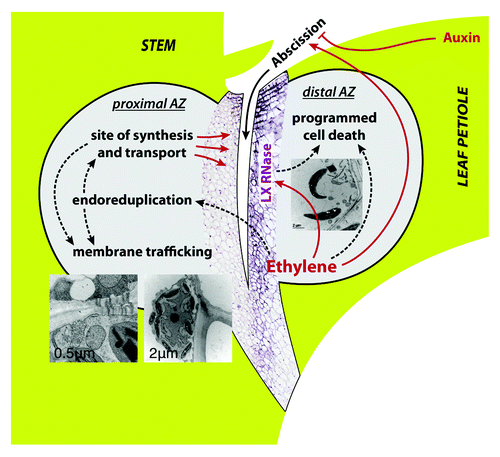
Figure 2. Endoreduplication in the AZ of untreated control (A) and debladed plant, 36 h after ethylene treatment (B). In situ DNA content of nuclei is shown as bubble graphs, superimposed over the images of Feulgen-stained median longitudinal tissue sections of AZ that were measured. For measuring the amount of nuclear DNA the interphase-peak method was applied.Citation9-Citation11,Citation19 The method adapted for use with tissue sections allows spatial and temporal examination of the distribution of nuclei. Nuclei of different endopolyploidy classes are color-coded in bubble graphs. Diameter of bubbles is linearly related to the diameter of measured nuclei. The site of future fracture is shown by dashed line in (A); fracture is delineated by black line in (B). The area visible in each panel is represented in the schematic drawing of a stem segment containing AZ in the lower left.
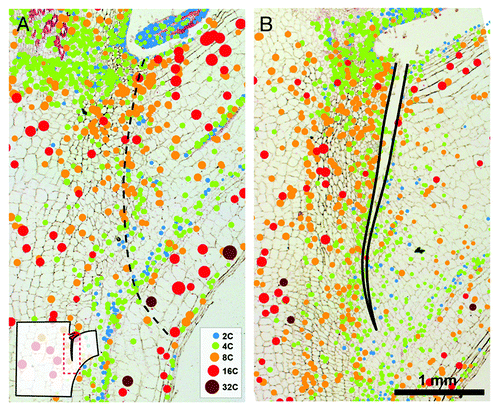
Figure 3. Cycle values of nuclei in proximal and distal side of AZs. The cycle value is calculated from the number of nuclei of each ploidy level multiplied by the number of endoreduplication cycles necessary to reach the corresponding ploidy level. The sum of the resulting products is divided by the total number of nuclei measured.Citation5 The cycle values were measured separately on proximal and distal side of AZs using nuclei distribution maps generated in ImageJ. Average cycle value and SD of five measurements per treatment are shown.