Figures & data
Figure 1. Northern blot analysis of decoy-targeted miRNAs of interest. A bulged (3nt) decoy targeting (A) miR171 and (B) miR319 was embedded in a long non-coding transcript and constitutively overexpressed in soybean, leading to decreased levels of each mature miRNA. (C) A bulged and mismatched decoy was each singly overexpressed in the same long non-coding transcript, each targeting miR160. Levels of mature miR160 were reduced in transformants containing each type of decoy, including one bulge event showing almost total elimination of the miRNA. (D) Levels of mature miR156 and miR172 were simultaneously reduced by overexpressing a double decoy transcript, a long non-coding transcript containing a bulged decoy site targeting miR156, followed by a second bulged decoy site targeting miR172. The decoy sites were separated by a non-coding 75 nucleotide spacer.
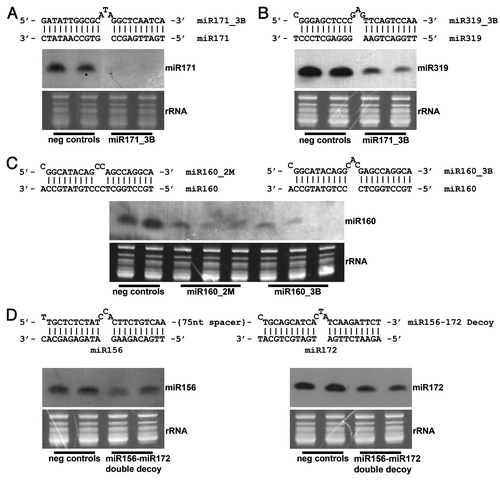
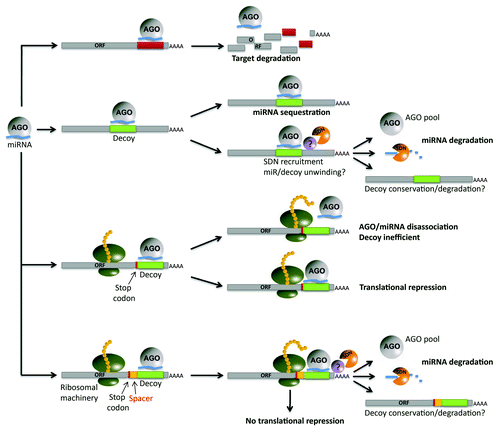
Table 1. Computational prediction of miRNA decoys in plant species representing important lineages
Figure 2. Potential miRNA interactions. Mature miRNAs loaded into AGO bind to and inhibit target transcripts through cleavage or via translational inhibition (top). One must take into consideration the interactions of miRNAs with decoys embedded in long non-coding transcripts (second). The miRNAs are potentially sequestered by the decoy, or alternatively, degraded, a process that requires the presence of SDNs in Arabidopsis. Little is known about the stability of the non-coding transcript after interaction with the miRNA has taken place. A third path is the interaction with a decoy site found within a coding transcript. If the decoy site is located too closely to the ORF, putative steric interactions between the ribosomal machinery and the miRNA may be possible. Translational repression is another consequence. If the decoy is located in the 3′ UTR at an adequate distance from the stop codon (bottom), the decoy is efficient and translation will not be affected.