Figures & data
Figure 1. Primary root growth analyses of CML9 genotype in reponse to flagellin treatment (A) Wild-type (Col), cml9 genotypes (KO cml9–1 and OE-CCs lines) and fls2 mutant seedlings grown for 7 d in MS medium (control, upper panel) in presence or not of flg22 1µM (lower panel). (B) Quantitative analyses of the primary root growth under control condition (MS) or after flagellin treatment (MS + flg22). The experiments were performed using three independent biological replicates and each histogram represents the mean root length (± SEM) analyzed using 20 to 24 independent roots per genotype. Statistical differences between the genotypes treated or not by flg22 are detected by ANOVA analysis followed by Tukey’s HSD test, at p < 0.05.
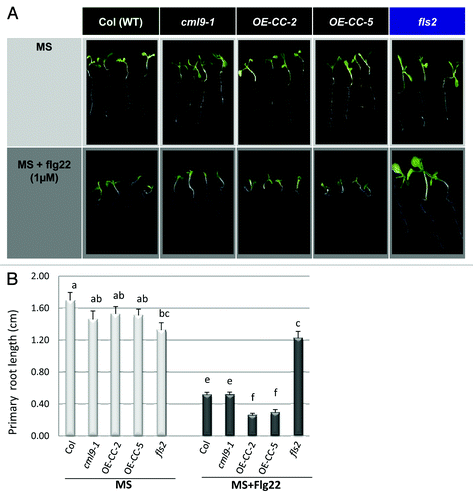
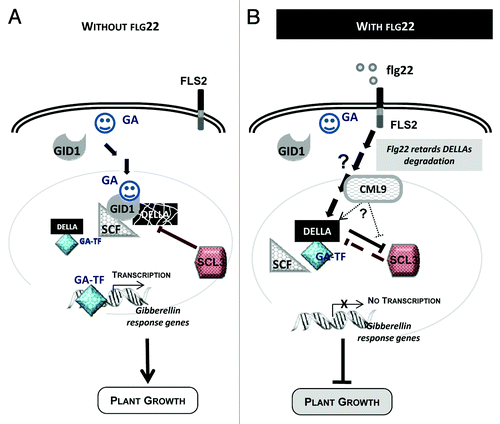
Figure 2. Hypothetical model involving CML9 in flg22-induced plant growth inhibition through GA-dependent signaling pathway. (A) Model of plant growth control through GA-dependent signaling. Upon GA binding to its soluble receptor (GID1), signaling pathway is activated; DELLA proteins are degraded via the ubiquitin-proteasome pathway. The major players of this GA-signaling cascade are the GA receptors (GID1), the DELLA repressor proteins and the F-box protein part of the SCF E3 Ubiquitin ligase complex. DELLAs are nuclear transcriptional regulators, which interact with other transcription factors (i.e. GA-TF, GA-dependent transcription factor) to modulate expression of GA-responsive genes. (B) Working model of flg22-induced plant growth inhibition that involve GA signaling cascade components and CML9.