Figures & data
Figure 1. Alternative Splicing of RVE8. (A) RVE8 (At3g09600) gene organization with exons depicted as rectangles, and introns as intervening horizontal lines. Protein coding exons/regions are shaded gray, UTR regions are depicted as clear rectangles. The region of RVE8 encoding the predicted single Myb domain is represented as hatched rectangles. Primer locations are denoted by arrows. Primer sequences are: RVE8#1-F GCTGGACAGAGGAAGAGCAC; RVE8#1-R TGCTCCACGAAGAGTAGCAA; RVE8#2-F GGGATATGCTTCATGGGATG; RVE8#2-R GCTGATTTGTCGCTTGTTGA. (B) Representative HR RT-PCR electropherograms for RVE8. AS isoforms were characterized from pooled RNAs representing plants harvested under both diurnal and constant light conditions at either normal ambient temperature or for plants undergoing cooling. Electropherograms show the size of detected peaks corresponding to RT-PCR products from alternatively spliced transcript variants. The x-axis is the size in bp and the y-axis represents arbitrary scales (relative fluorescent units) to reflect relative abundance of peaks. Individual products are indicated and AS variants are described in more detail in . FS – fully spliced product; ASn – alternative splicing variant where n is the number to identify the product. AS transcripts were detected on an ABI3730 automatic DNA sequencer along with GeneScan™ 1200 LIZ size standard. Amplicons were accurately sized using GeneMapper software. For more details of Methods seeCitation15
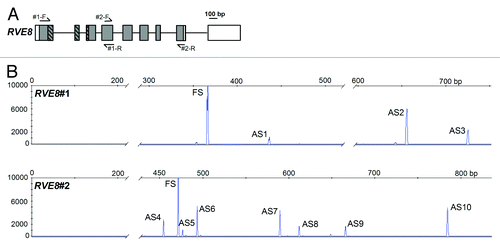
Table 1. Alternative splicing events in RVE8
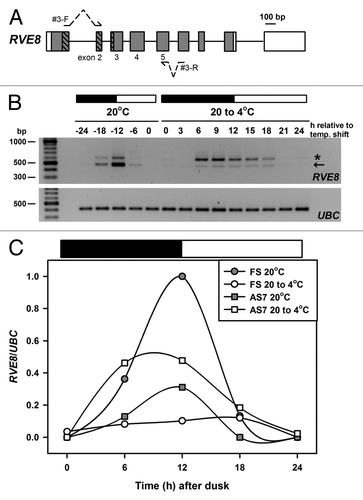
Figure 2. Temperature associated AS of RVE8. (A) RVE8 gene organization is as outlined in . To preclude potential amplification of gDNA during RT-PCR both forward and reverse primers – shown as arrows interrupted with dotted lines – spanned an intron and had the potential to amplify AS transcripts between exons 2 and 5. Primer sequences are: RVE8#3-F; TGAAGCACTTCAACTGTTTGATCGTGACTG and RVE8#3-R; ATCAGGAACACCGTGAAGCACTGGA. Bases in bold are separated by an intron in the RVE8 gDNA sequence. (B) RT-PCR amplification of RVE8, using the primer sequences described above, for plants harvested during diurnal cycles of 12 h dark and 12 h light (denoted by the dark and white bars, respectively) either at ambient temperature (20°C) or during a low temperature transition (20 to 4°C). The cooling temperature shift was initiated at dusk at 20°C and samples are labeled as relative to the temperature shift. Arrow denotes the fully spliced or canonical RT-PCR product (predicted size 481 bp), and the star an AS product, representing transcripts retaining intron 4 (AS7, predicted size 599 bp, see also and ). Note that samples are every 6 h at 20°C and every 3 h during the cold transition. For more details of Methods, and for the UBC primer sequences, seeCitation15 (C) RVE8 FS and AS7 RT-PCR band intensities (normalized to the UBC positive controls) from panel B were plotted across the diurnal cycles of 12 h dark and 12 h light (denoted by the dark and white bars, respectively) either at ambient temperature (20°C) or during a low temperature transition (20 to 4°C). Band intensities were determined using a BioRad GelDoc 2000 imaging system and QuantityOne software.