Figures & data
Figure 1. Ligand specificity of MAMP-stimulated iGluRs and molecular control of iGluR desensitization.10–12-d-old seedlings were pre-treated for 30 min with (co-) agonists, W-7 or CHX as described in Kwaaitaal et al. (2011) and then 1 μM flg22 was added to trigger a Ca2+ transient. Seedlings were pre-treated with (A, B) 1 mM NMDA, 1 mM d-serine, 1 mM L-glutamate, (C, D) 1 mM AMPA, 1 mM kainic acid, (E, F) 50 μM W-5 or W-7 or (G, H) 100 μM CHX. Shown are the absolute Ca2+ concentrations (A, C, E, G) or the background-corrected differences in cellular Ca2+ levels (B, D, F, H). Data given are the mean ± standard deviation of 6–12 seedlings per measurement and represent typical examples of three or more independent experiments. Note that the first short and strong Ca2+ peak marked by the gray bar results from the mechanical stimulation caused by the addition of the stimulus. Also note the large standard deviation of the W-7-treated seedlings compared with the control measurements, which reflects the high variation in the individual seedling curves upon treatment with this compound (see Fig. S2). For further experimental details see reference Citation3.
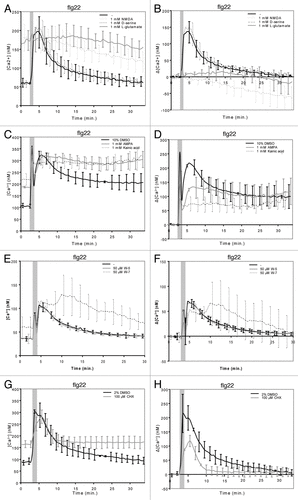
Figure 2. Presence of a potential CaMBD at the cytoplasmic C-terminus of Arabidopsis iGluRs. Transmembrane domain prediction was performed using Aramemnon (http://aramemnon.botanik.uni-koeln.de/) website and the stretches predicted using a hidden Markov modelCitation29 were marked. Putative CaMBDs (marked in gray) were identified based on the following web tool: http://calcium.uhnres.utoronto.ca/ctdb/ctdb/sequence.html.Citation23 For the prediction the last 268–375 amino acids were used, starting from the first TM domain and excluding the extracellular N-terminal domain. Marked residues represent those having a score higher than 6. Alignments were generated in the CLC Workbench package (www.clcbio.com).
