Figures & data
Figure 1. Phylogenetic relationship of FAH. A phylogenetic tree of FAH proteins derived from plants, animals and fungi was constructed by the CLUSTAL W program. Protein sequences were obtained from the National Center for Biotechnology Information. AtFAH1, At2g34770; AtFAH2, At4g20870; Oryza sativa 1, Os12g0628400; Oryza sativa 2, Os03g0780800; Glycine max 1, ACU24407; Glycine max 2, ACU24572; Sorghum bicolor 1, XP_002466355; Sorghum bicolor 2, XP_002442581; Vitis vinifera 1, XP_002268031; Vitis vinifera 2, XP_002267991; Homo sapiens, NP_077282; Mus musculus, NP_835187; Rattus norvegicus, NP_001129055; Gallus gallus, XP_414053; Xenopus laevis, NP_001082707; Nematostella vectensis, XP_001639353; Ciona intestinalis, XP_002126665; Caenorhabditis elegans, NP_492678; Drosophila ananassae, XP_001959617; Kluyveromyces lactis, XP_453142; Lachancea thermotolerans, XP_002552404; Saccharomyces cerevisiae, NP_013999; Candida glabrata, XP_446117; Ashbya gossypii, NP_982359; Pichia pastoris, XP_002492764; Yarrowia lipolytica, XP_505342; Magnaporthe oryzaeitalic>, XP_361446; Neosartorya fischeri, XP_955879; Aspergillus flavus, XP_002373274; Aspergillus oryzae, XP_001818088; Neosartorya fischeri, XP_001264167; Aspergillus fumigatus, XP_752948; Penicillium chrysogenum, XP_002561746.
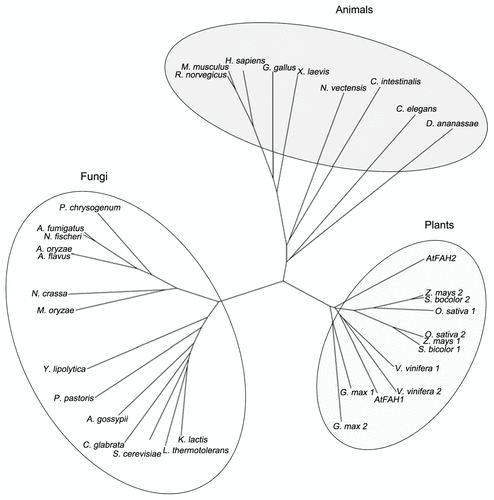
Figure 2. Comparison of deduced amino acid sequences of FAH. Amino acid sequences were derived from Arabidopsis thaliana (AtFAH1, AtFAH2), Oryza sativa, Glycine max, Zea mays, Sorghum bicolor, Vitis vinifera, Homo sapiens, Gallus gallus, Xenopus laevis, Drosophila ananassae, Caenorhabditis elegans, Saccharomyces cerevisiae, Neurospora crassa and Aspergillus oryzae. Dark gray indicates HPGG, which is one of the most important motifs in the Cb5 protein family, and light gray indicates the histidine motif.
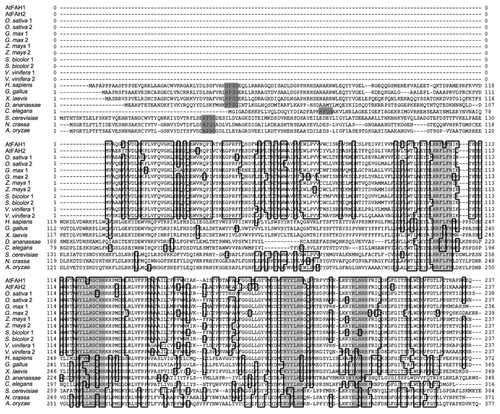
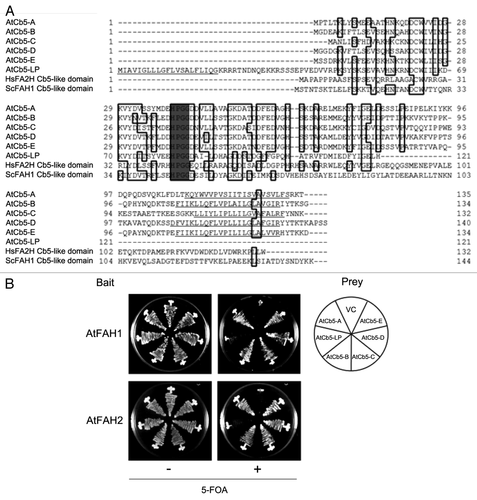
Figure 3. Interactions of AtFAHs with AtCb5 family proteins. (A) Comparison of deduced amino acid sequences among AtCb5 family proteins (AtCb5-A, -B, -C, -D, -E and -LP), and Cb5-like domains of human FA2H (HsFA2H) and yeast FAH1 (ScFAH1). Gray indicates HPGG. Possible transmembrane domains predicted by SOSUI WWW Server are underlined. (B) Interactions between AtFAHs and AtCb5 family proteins were detected by the suY2H system. Yeast cells containing bait and prey constructs were tested for their binding on minimum medium containing 5-fluoroorotic acid (5-FOA). The results indicate the growth of each line after 2 d (without 5-FOA) and 3 d (with 5-FOA) of incubation at 30°C. VC indicates the combination of vector controls.