Figures & data
Figure 1. Sequence alignment of the Arabidopsis CRT3 and its plant homologs. Amino acid sequence alignment of Arabidopsis CRT3 and its plant homologs was performed using the ClustalW program at Mobyle web tool (http://mobyle.pasteur.fr/cgi-bin/portal.py). Names and accession numbers of the CRT3 homologs used for analyses are as follows: AtCRT3 (NP_563816; At: Arabidopsis thaliana), BrCRT3 (translated from nucleotide sequences EX116073.1, EX026998.1 and EX117522.1, Br: Brassica rapa), VvCRT3 (translated from nucleotide sequence XM_002276397.1, Vv: Vitis vinifera), MtCRT3 (translated from nucleotide sequence BT052978.1, Mt: Medicago truncatula), OsCRT3 (BAC06263; Os: Oryza sativa), GhCRT3 (translated from nucleotide sequence DT563804.1; Gh: Gossypium hirsutum) MdCRT3 (translated from nucleotide sequences CN943087.1 and CV082227.1; Md: Malus domestica), SmCRT3 (translated from nucleotide sequence FE490612.1 and FE490611.1; Sm: Selaginella moellendorffii), AfCRT3 (translated from nucleotide sequences DR940519.1 and DT750168.1, Af: Aquilegia formosa), PtCRT3 (translated from nucleotide sequences BF777977.1, CO198952.1 and CO158387.1; Pt: Pinus taeda), ZmCRT3 (translated from nucleotide sequence AY105822; Zm: Zea mays), HvCRT3 (translated from nucleotide sequence AK248906.1; Hv: Hordeum vulgare) and TaCRT3 (EF452301.1; Ta: Triticum aestivum). Aligned amino acid sequences were shaded using the BoxShade 3.31 web server (http://mobyle.pasteur.fr/cgi-bin/portal.py). Residues identical in more than 8 sequences are shaded in red and similar ones are shaded in cyan. Two basic tetrapeptides in the C terminus of AtCRT3 are marked by blue bar, and the region that has > 50% basic residues is indicated by a green box.
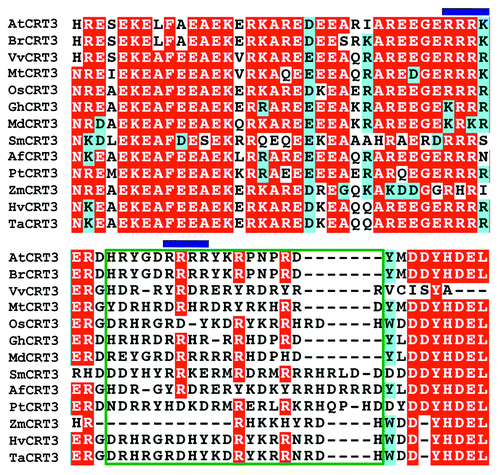
Figure 2. Mutations of the basic Arg392Arg393Arg394Lys395 tetrapeptide destroy the activity of the gCRT3M transgene to rescue a T-DNA insertional ebs2 mutation. (A) Images of 5-week-old soil-grown plants of wild-type (WT) plant, bri1-9, ebs2-8 bri1-9 and 4 representative gCRT3M ebs2-8 bri1-9 transgenic lines. (B) Immunoblot analysis of CRT3M abundance in transgenic plants. Total protein extracts of 5-week-old soil-grown plants shown in (A) were separated by 10% SDS/PAGE and analyzed by immunoblotting with anti-CRT3 antibody. Coomassie blue staining of a small subunit of the Arabidopsis Rubisco gel was used as a loading control. The protein samples of 4 chosen gCRT3M ebs2-8 bri1-9 transgenic plants were diluted 2 fold before sample loading.
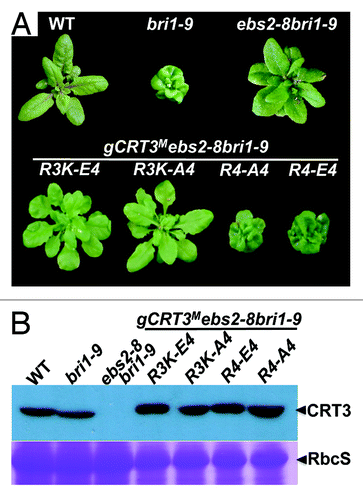
Figure 3. The Endo-H sensitivity assay of BRI1 and bri1-9. Equal amounts of total proteins extracted from leaves of 5-week-old soil-grown plants shown in were treated with (+) or without (−) Endo Hf for 1.5 h at 37°C, separated by 7% SDS/PAGE and analyzed by immunoblotting with an anti-BRI1 antibody. Coomassie blue staining of RbcS serves as a loading control. Arrows indicated the positions of Endo-H-resistant (bri1-9R) and Endo-H-sensitive bri1-9 (bri1-9S) forms.
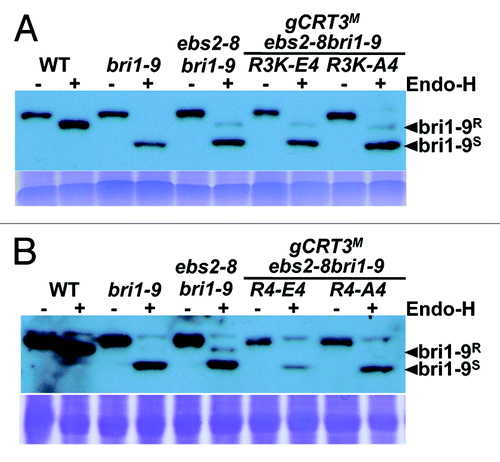
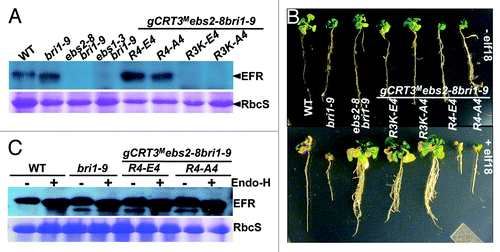
Figure 4. The basic tetrapeptide Arg392Arg393Arg394Lys395 of CRT3 is crucial for the complete folding of the plant immunity receptor EFR. (A) An immunoblot analysis of EFR. Total protein extracts from WT, bri1-9, ebs2-8bri1-9 and 4 selected gCRT3M ebs2-8 bri1-9 transgenic lines were separated by 10% SDS/PAGE and analyzed by immunoblotting with an anti-EFR antibody. Coomassie blue staining of RbcS serves as a loading control. (B) The efl18 growth inhibition assay. Shown here are images of 7-d-old seedlings of WT, bri1-9, ebs2-8bri1-9 and 4 selected gCRT3M ebs2-8 bri1-9 transgenic lines grown on half-strength MS agar medium (top panel) and 17-d-old seedlings of the same genotypes after 10-d treatment with 100 nM elf18 (bottom panel). (C) The Endo-H assay of EFR. Equal amounts of total proteins extracted from leaves of 5-week-old soil-grown plants were treated with (+) or without (−) Endo Hf for 1.5 h at 37°C, separated by 7% SDS/PAGE and analyzed by immunoblotting with an anti-EFR antibody. Coomassie blue staining of RbcS serves as a loading control.