Figures & data
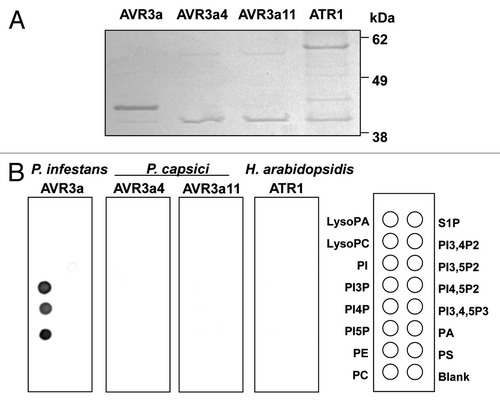
Figure 1. The NMR-derived model structure of AVR3a with the RXLR motif. The residues for the RXLR motif (R44, L45, L46 and R47) and PIP binding (K85) are mapped on the ribbon diagram as red sticks.
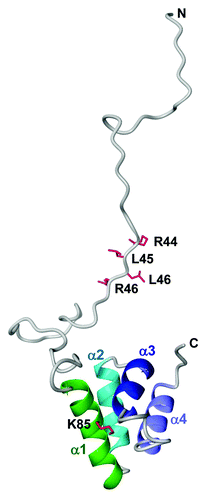
Figure 2. Lipid overlay assay of oomycete RXLR effectors, AVR3a, AVR3a4, AVR3a11 and ATR1. (A) Escherichia coli strain BL21-AI was transformed with pDEST24 constructs for AVR3a (Asp23-Tyr147), AVR3a4 (Asn22-Tyr122), AVR3a11 (Asn22-Val132) or ATR1-Emwa1 (Ser22-Glu324). Protein expression and purification were performed as described in Yaeno et al.Citation5 The purified C-terminal GST fusion proteins were checked by SDS-PAGE stained with InstantBlue (Expedeon) and equal amounts of proteins were used for the lipid overlay assay. (B) Nitrocellulose membranes spotted with 100 pmol of various lipids (PIP Strips; Echelon Biosciences) were blocked in 1% nonfat milk in PBS for 1 h and then incubated with 1 μg/mL C-terminal GST fusions of P. infestans AVR3a, P. capsici AVR3a4, P. capsici AVR3a11 and H. arabidopsidis ATR1 overnight at 4°C. After washing with PBS-T, the bound proteins were detected using anti–GST-HRP antibodies (GE Healthcare) diluted to 1:2,000. PA, phosphatidic acid; PC, phosphatidyl-choline; PE, phosphatidylethanolamine; PI, phosphatidylinositol; PI3P, PI-3-phosphate; PI4P, PI-4-phosphate; PI5P, PI-5-phosphate; PI3,4P2, PI-3,4-biphosphate; PI3,5P2, PI-3,5-biphosphate; PI4,5P2, PI-4,5-biphosphate; PI3,4,5P3, PI-3,4,5-triphosphate; PS, phosphatidylserine; S1P, sphingosine-1-phosphate.