Figures & data
Figure 1. Estimation of chlorophyll index (SPAD-values), leaf area index (LAI) and proline content in wheat genotypes viz., DBW-140, RAJ-3765, PBW-574, K-0-307 and HS-240 under timely and late sown conditions. Leaf chlorophyll content (SPAD-values) was recorded in the flag leaves, using a self-calibrating SPAD chlorophyll meter (Minolta). Thirty flag leaves per plant were used to calculate the chlorophyll index value that is proportional to the amount of chlorophyll (A). Leaf area index (LAI) was measured with the help of Plant Canopy Analyzer (LAI-2000, LI-COR) (B). Proline content (μg g−1 FW) in wheat genotypes was calculated at anthesis and at 15 DAS (days after anthesis) (C). Vertical lines on top of bars indicate standard error of means.
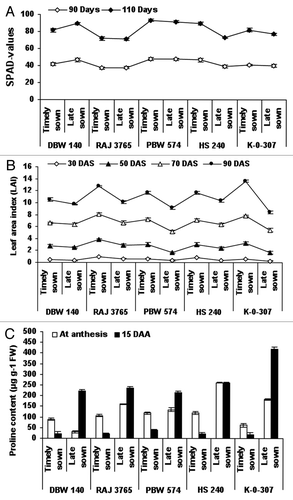
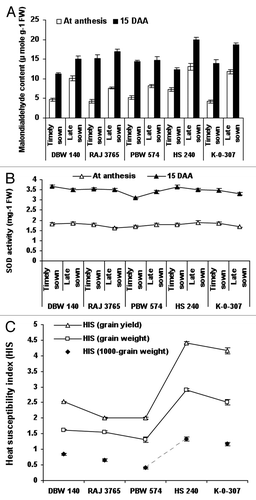
Figure 2. Evaluation of malondialdehyde (MDA) content, superoxide dismutase (SOD) activity and heat susceptibility index (s) (HIS) in wheat genotypes viz., DBW-140, RAJ-3765, PBW-574, K-0-307 and HS-240 under timely and late sown conditions. The extent of lipid peroxidation was evaluated by calculating the concentration of thiobarbituric acid reactive substances as malondialdehyde equivalent using the extinction coefficient (155 mM−1 cm−1) (A). Superoxide dismutase activity was determined by measuring its ability to inhibit the photochemical reduction of nitro-blue tetrazolium (NBT) in the presence of riboflavin in light (B). The heat susceptibility index (s) for yield characters per genotype was calculated (C). Vertical lines on top of bars indicate standard error of means.