Figures & data
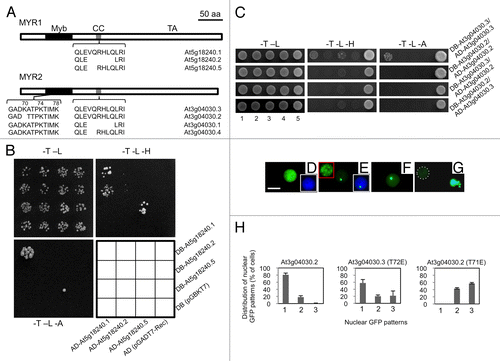
Figure 1. Alternative splicing of MYR1 and MYR2 affects dimerization and localizations (A) Schematic diagram of alternative splicing in MYR1 and MYR2. Both MYR1 and MYR2 contain 3 domains: Myb-like DNA-binding domain (Myb), coiled-coil domain (CC), and transactivation domain (TA). Conserved alternative splicing at the CC domain of MYR1 and MYR2 results in 3 isoforms. In addition, alternative splicing at the Myb-like DNA-binding domain of MYR2 yields 2 isoforms. (B) MYR1 isoforms produced from alternative splicing at the CC domain form 2 distinct homodimers in yeast and possibly one heterodimer. Yeast AH109 cells co-transformed with 2 plasmid DNAs as shown in the interaction grid were spotted on SD plates lacking tryptophan and leucine (-T -L), tryptophan, leucine, and histidine (-T -L -H), or tryptophan, leucine, and adenine (-T -L -A), and incubated at 30 °C for 3–10 d. (C) MYR2 isoform At3g04030.2, produced from alternative splicing at the Myb domain, forms neither a homodimer nor a heterodimer with At3g04030.2 or At3g04030.3, respectively, while At3g04030.3 can form a homodimer. For different MYR2 isoform assays (C, at right) yeast AH109 was co-transformed with 2 plasmid DNAs, as exemplified by the DB-At3g04030.3/AD-At3g04030.3 assay using plasmid combinations 1 through 5: 1, DB-At3g04030.3, AD-At3g04030.3; 2, DB-At3g04030.3, AD; 3, DB, AD-At3g04030.3; 4, DB, AD; 5, positive control (pGBKT7–53 + pGADT7-RecT), and grown on drop-out plates as shown. (D-H) Subcellular localization of GFP-MYR2 isoforms and mutants transiently expressed in Nicotiana benthamiana. MYR2 isoform GFP-At3g04030.3 was evenly distributed throughout the nucleus in all cells (D). In most cells (79%), GFP-At3g04030.2 was also evenly distributed throughout the nucleus. However, in 21% of cells (n = 1771), a portion of GFP-At3g04030.2 localized to a single spot or multiple spots (inset in red box) in the nucleus (E and H, left panel). Insets in white boxes (D, E) show GFP/DAPI overlay images for nuclei. Point mutants K70A (F) or K74A (not shown) of GFP-At3g04030.3 mimic the nuclear spot localization pattern seen for GFP-At3g04030.2. The double mutant (K70A K74A, K70A K78A, and K74A K78A) forms of GFP-At3g04030.3 aggregate outside of the nucleus in some cells, while triple mutant (K70A K74A K78A) GFP-At3g04030.3 forms extra-nuclear aggregates in all GFP-positive cells (G). Nucleus in (G) is surrounded by dashed white line. T72E (At3g04030.3) (H, center panel; n = 445) or T71E (At3g04030.2) (H, right panel; n = 715) mutations enhanced the localization of GFP to one or multiple spots in the nucleus. Categories of GFP localization (H) are: 1, evenly distributed throughout the nucleus; 2, localized to a single nuclear spot; 3, localized to multiple nuclear spots. Bar in (D) = 10 µm for (D-G).