Figures & data
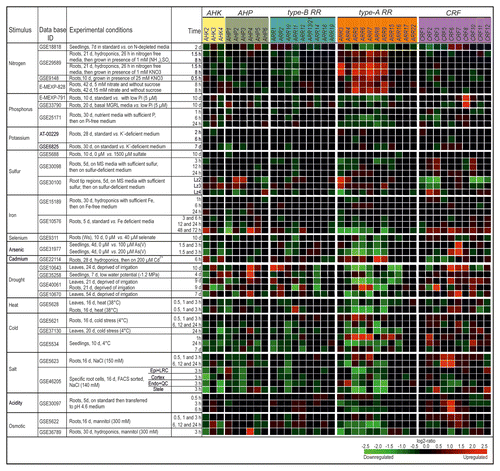
Figure 1. Regulation of CK metabolism genes by different environmental cues. The changes in transcript abundance of the CK metabolism genes in response to different environmental cues or stresses were established with the Genevestigator databaseCitation11 (http://www.genevestigator.com/gv/index.jsp) and modified. The figure shows an arbitrary selection of experiments according to criteria outlined in the text. The gene expression responses were calculated as log2-ratios between the signal intensities from different stress or nutritional treatments compared with control or mock-treated samples. The resulting heatmap is color-coded as indicated and thus reflects the up- (red color) or downregulation (green color) of genes. The individual experiments are available from various repositories, such as Gene Expression OmnibusCitation25 or ArrayExpressCitation26 and can be retrieved by using their unique ID. Note that a probe for CKX7 is not present on the Affymetrix ATH1 chip. Abbreviations: d, days; vs., versus; Lz, longitudinal zone; Ws, Wassileskija; Col-4, Columbia-4.
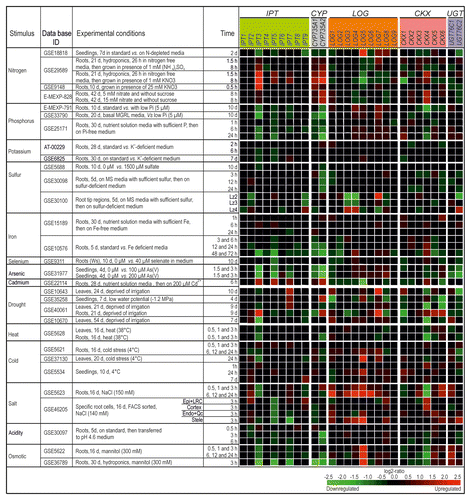
Figure 2. Regulation of CK signaling genes by different environmental cues. The changes in transcript abundance of CK signaling genes in response to different environmental cues or stresses was established with the Genevestigator databaseCitation11 (http://www.genevestigator.com/gv/index.jsp) and modified. The figure shows an arbitrary selection of experiments as is outlined in the text. The gene expression responses were calculated as log2-ratios between the signal intensities from different stress or nutritional treatments compared with control or mock-treated samples. The resulting heatmap is color coded as indicated and thus reflects the up- (red color) or downregulation (green color) of genes. The individual experiments are available from various repositories, such as Gene Expression OmnibusCitation25 or ArrayExpressCitation26 and can be retrieved by using their unique ID. Please note that only a single probe has been annotated on the Affymetrix ATH1 chip for both ARR13 and ARR21 and probes for CRF4 and CRF9 were not present. Abbreviations: d, days; vs., versus; Lz, longitudinal zone; Ws, Wassileskija; Col-4, Columbia-4.