Figures & data
Figure 1. CDPKs from Chlamydomonas reinhardtii. (A) Schematic diagram of CDPK1 and CDPK3 showing the C2 domain from 1–95 aa in CDPK1, protein kinase catalytic domains (STK), and the 4 EF hands in both the CDPKs. (B) Phylogenetic relationship between C. reinhardtii CDPK1, CDPK3, and other CDPKs from algae, moss, higher plants, and apicomplexans. The phylogenetic tree was inferred using the Neighbor-Joining method. The optimal tree with the sum of branch length = 4.60159844 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths (next to the branches) in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. The analysis involved 20 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 371 positions in the final data set. Evolutionary analyses were conducted in MEGA6.
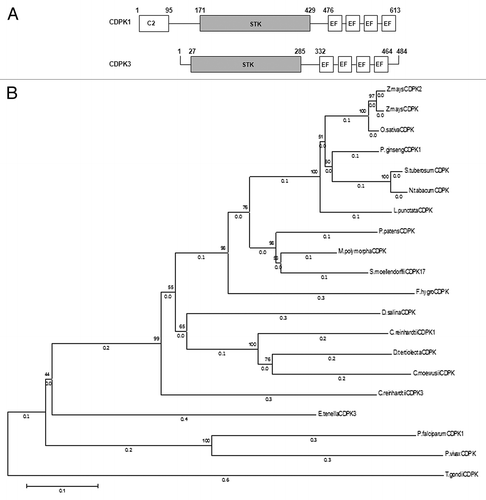
Figure 2. Expression patterns of CDPK1 and CDPK3 transcripts in response to nutrient starvation at 0, 1, 3, 6, and 18 h by real-time RT-PCR analysis. (A) Acetate starvation. (B) Phosphorus starvation. (C) Nitrogen starvation. The single and double asterisks indicate a statistically significant difference at P< 0.05, P< 0.005 respectively.
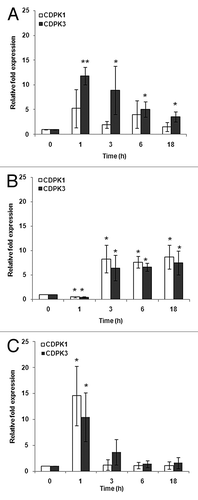
Figure 3. Ca2+ mobility of C. reinhardtii CDPK1-GFP. (A) CDPK1-GFP construct used to transform C. reinhardtii cells and (B) Ca2+ mobility shift of CDPK1-GFP fusion protein. Lanes 1, 2, 4, and 5 are treatments with different concentrations of CaCl2; lane 3 is the lysate treated with EGTA only. CDPK1-GFP cell lysate treated with CaCl2 (10 mM, 5 mM, 2.5 mM, and 1 mM) and EGTA (2.5 mM) were run on a 7% SDS -PAGE followed by Western Blotting using anti-GFP antibody. CDPK1 showed a mobility shift in presence of Ca2+.
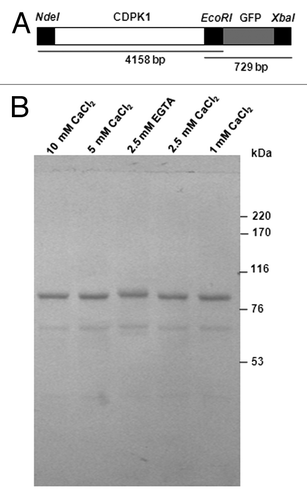
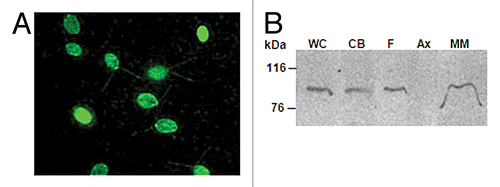
Figure 4. C. reinhardtii CDPK1 is localized in the cell body and flagella. (A) Immunostaining of cells expressing CDPK1-GFP with anti-GFP antibody. CDPK1-GFP fusion protein was localized in C. reinhardtii flagella and cell body. (B) Whole cell (WC), cell body (CB), isolated flagella (F), axoneme (Ax), and membrane-matrix (M-M) fractions of CDPK1-GFP clone were analyzed by immunoblotting using anti-GFP antibody. CDPK1- GFP fusion protein was present in the cell body and the membrane-matrix fraction of the flagella. The bar on (A) indicate 10 µm.