Figures & data
Figure 1. Impact of heat stress on the number and the size of leaves in the progeny of heat stressed and control plants. Plants were stressed at 50 °C for 3 h, on 5 consecutive days. Measurements were taken for leaf number, length, and width. All confidence intervals are a result of bootstrap x 10,000, at 95% confidence (P = 0.05) (A-C) Leaf number (A), length (B), and width (C) in F1 progeny of wild type (15D8) and mutant (dcl2, dcl3, and dcl4) plants, as indicated by Legend, exposed to heat and control plants. “C1” – the progeny of plants grown at normal conditions in F0. “S1” – the progeny of plants exposed to heat in F0. “+” and “-” indicate exposure to stress or growth in uninduced conditions, respectively, each of which had approximately 24 plants. Asterisks (*) indicate a significant difference from wild type, with the same treatment group, or difference between treated and non-treated plants within the same F0 treatment (C1+ vs C1- and S1+ vs S1-) within the same mutant, as calculated using a t test (P ≤ 0.05). (D) Bars represent ratio of change in F1 plants in response to heat stress (S+/S- or C+/C-), and those that overlap with value of 1 indicate no significant change under heat stress. Asterisks (*) are used to indicate mutants that are significantly different from wild-type (15D8) plants with the same treatment. Legend indicates parental treatment.
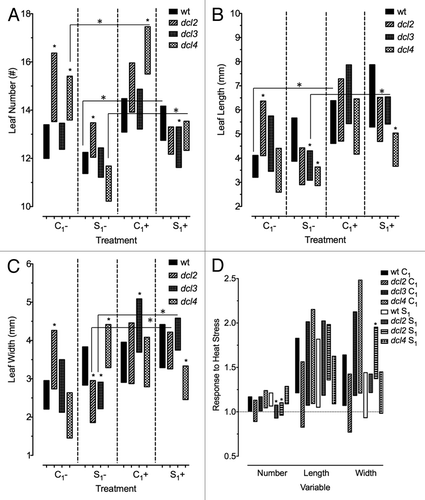
Figure 2. Size of seeds produced by heat stressed and control plants. Approximately 100–200 seeds per sample group. (A) Seed size in F1 progeny of wild type (15D8) and mutant (dcl2, dcl3, and dcl4) plants, as indicated by Legend, exposed to heat and control plants. “C1” – the progeny of plants grown at normal conditions in F0. “S1” – the progeny of plants exposed to heat in F0. Asterisks (*) indicate a significant difference from wild type, with the same treatment group, or difference between different treatments (C1 vs S1) within the same mutant, as calculated using a t test (P ≤ 0.05). (B) Bars represent ratio of change in seeds produced by plants that had been heat stressed (S1/C1), and those that overlap with value of 1 indicate no significant difference in seed size in response to heat stress. Asterisks (*) are used to indicate mutants that are significantly different from wild-type (15D8) plants with the same treatment, as calculated using a t test (P ≤ 0.05). Legend indicates parental treatment.
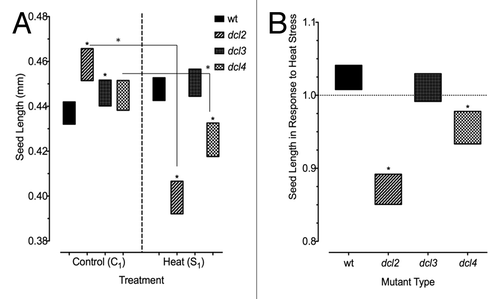
Figure 3. Percentage of F1 plants that had bolted at approximately 4 wk of age. Approximately 24 plants were included in each treatment group. (A) Comparison of bolting in F1 plants when grown at normal conditions. (B) Comparison of bolting in F1 plants when grown under either normal (-) or stressed (+) conditions. Each treatment group is labeled according to treatment and mutant type, on the horizontal axis.
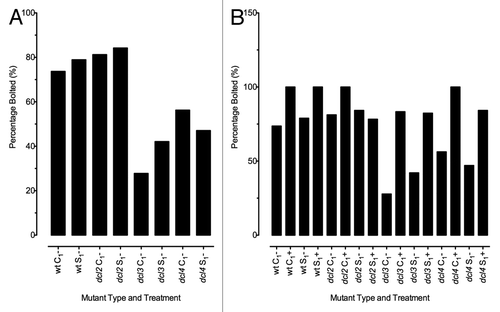
Figure 4. Expression of ONSEN (A) and TSI (B) transposons in F1 progeny of wild type (15D8) and mutant (dcl2, dcl3, and dcl4) plants exposed to heat and control plants. Y-axis shows arbitrary units of gene expression. “C1” – the progeny of plants grown at normal conditions in F0. “S1” – the progeny of plants exposed to heat in F0. “+” and “-” indicate exposure to stress or growth in uninduced conditions, respectively. Bars show standard deviation calculated from 3 technical repeats. Asterisks (*) indicate a significant difference between control (-) and stress (+) with the same parental treatment or between different treatments (C1 vs S1), as calculated using a t test (P ≤ 0.05).
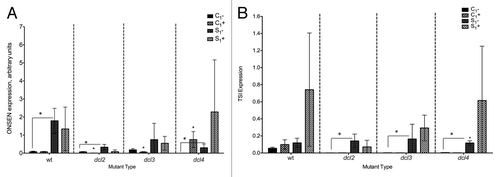
Figure 5. Analysis of global genome methylation in the F1 progeny of wild type (15D8) and mutant (dcl2 and dcl3) plants exposed to heat and control plants. Y-axis shows units of radioactive incorporation (dpm/µg). Higher incorporation indicates a lower degree of methylation. “C1” – the progeny of plants grown at normal conditions in F0. “S1” – the progeny of plants exposed to heat in F0. “+” and “-” indicate exposure to stress or growth in uninduced conditions, respectively. Error bars show standard deviation calculated from 2 technical repeats. Asterisks (*, **) indicate a significant difference between control (-) and stress (+) with the same parental treatment, between different treatments (C1 vs S1) or between different mutants, as calculated using a t test (P ≤ 0.05 or P ≤ 0.01, for 1 or 2 asterisks, respectively).
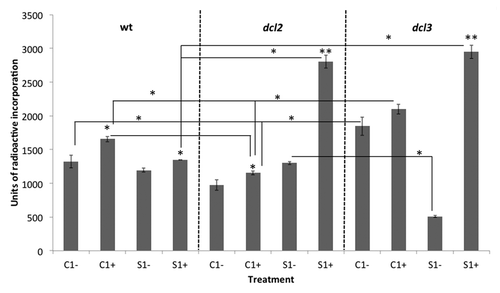
Figure 6. Expression of (A) HSFA2 (B) MSH6 (C) ROS1 in F1 progeny of wild type (15D8) and mutant (dcl2 and dcl3) plants exposed to heat and control plants. Y-axis shows arbitrary units of gene expression, standardized to tubulin. “C1” – the progeny of plants grown at normal conditions in F0. “S1” – the progeny of plants exposed to heat in F0. “+” and “-” indicate exposure to stress or growth in uninduced conditions, respectively. Bars show standard error of the mean (SEM) calculated from 3 technical repeats. Asterisks (*) indicate a significant difference between control (-) and stress (+) with the same parental treatment, between different treatments (C1 vs S1) or between different mutants, as calculated using a t test (P ≤ 0.05).
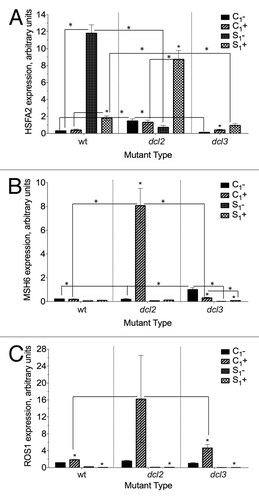
Figure 7. Expression of (A) SUVH2 (B) SUVH5 (C) SUVH6 (D) SUVH8 in F1 progeny of wild type (15D8) and mutant (dcl2 and dcl3) plants exposed to heat and control plants. Y-axis shows arbitrary units of gene expression, standardized to tubulin. “C1” – the progeny of plants grown at normal conditions in F0. “S1” – the progeny of plants exposed to heat in F0. “+” and “-” indicate exposure to stress or growth in uninduced conditions, respectively. Bars show standard error of the mean (SEM) calculated from 3 technical repeats. Asterisks (*) indicate a significant difference between control (-) and stress (+) with the same parental treatment, between different treatments (C1 vs S1) or between different mutants, as calculated using a t test (P ≤ 0.05).
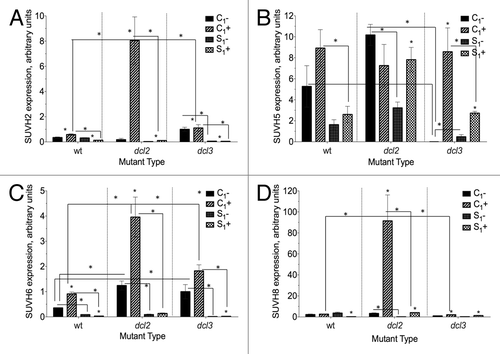
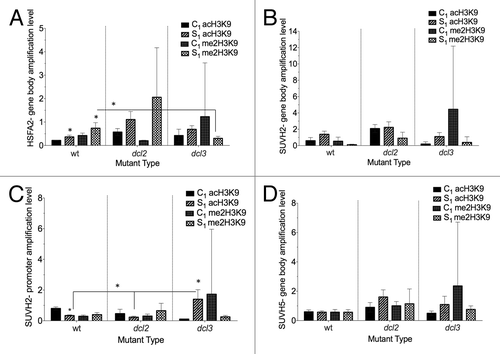
Figure 8. Analysis of H3K9me2 and H3K9ac histone modifications in HSFA2, SUVH2, SUVH5 genes. PCR analysis was done in the progeny of heat-stressed (S1) and control (C1) plants using primers specific to the gene body region of HSFA2 (A), promoter region of SUVH2 (B), gene body region of SUVH2 (C), and gene body region of SUVH5 (D). The y-axis shows the levels of H3K9me2/H3K9ac expression in average arbitrary units (calculated from 2 technical repeats). The x-axis indicates the mutant type. The asterisks (*) denotes a significant differences between the progeny of control and the progeny of stressed or between mutants for the same treatment, as determined with a t test (P ≤ 0.05).