Figures & data
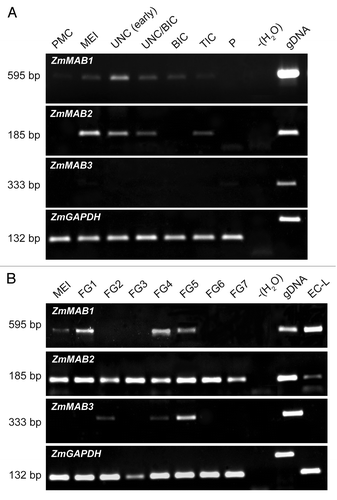
Figure 1. Unrooted phylogentic tree of 259 genes encoding MATH-BTB proteins from 9 representative land plant species (embryophyta). Analysis was done with Seaview v.4.3.4. using nucleotide sequence regions (open reading frames) of full-length MATH-BTB proteins with both MATH and BTB domains. Genes are color-coded by species (see legend for colors at the bottom right side of the tree). E1 to E5 indicate 5 major grass-specific subclades of the expanded clade. Core clade MATH-BTB genes form a separate clade. The number after decimal point for designated gene presents a splicing variant used for phylogenetic analysis. The bar of 0.5 is a branch length, which represents nucleotide substitutions per site. For sequence identifiers see Table S1. For statistical support of phylogenetic tree see Figure S1.
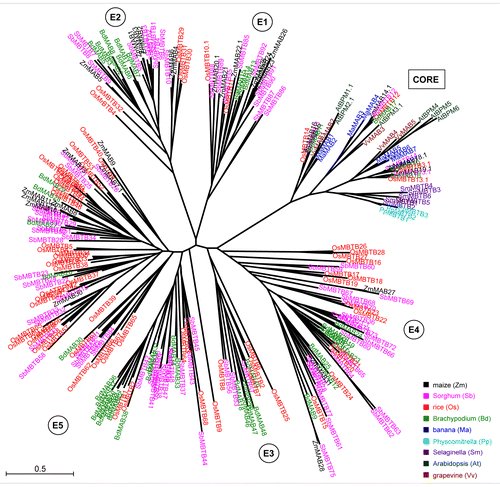
Figure 2. Graphic display of shared synteny and tandem duplications of selected grass MATH-BTB genes from the expanded subclades E1-E5 of rice, Brachypodium, Sorghum, and maize. MATH-BTB genes are depicted with the label MB, BTB genes with B, MATH domain encoded genes with M, and known MATH-BTB pseudogenes in rice with P. These genes are additionally highlighted in red. Selected members of the MATH-BTB family are colored in gray and appear in the center of the diagram surrounded by 5 upstream and downstream neighbor genes. Note that some MATH-BTB genes identified in this study are depicted in other colors. The graph was generated using the Phytozome database. To simplify visualization of genes from different species belonging to the same family other than MATH-BTBs, they were labeled with numbers and appear in the same color. For corresponding sequence identifiers matching labels see Table S2.
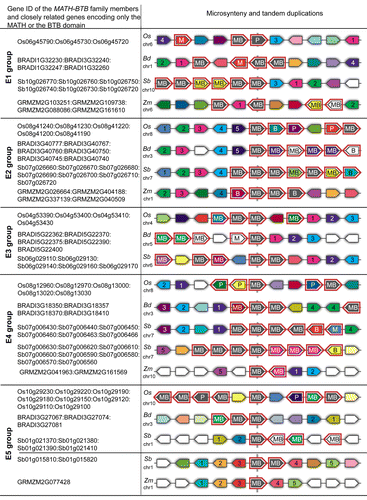
Figure 3. Expression of ZmMAB1–3 during germline developmental in maize inbred line A188. (A) Expression during male gametophyte development from pollen mother cell (PMC), meiosis (MEI), uninucleate microspore (UNC), bicellular pollen (BIC), tricellular pollen (TIC), and mature pollen at shedding phase (P; anthesis). (B) Expression during female gametophyte development from meiosis (MEI) to mature female gametophytes (stage FG7). Stages after Evans and Grossniklaus.Citation21 An egg cell library (EC-L;Citation22) was included as a positive control for ZmMAB1. RT-PCR with water (-H20) served as negative, and genomic DNA (gDNA) as a positive control for all genes. Maize GAPDH was used as a housekeeping control.