Figures & data
Figure 1. itt3 vacuoles are independent organelles. (A–B) Root epidermal cells from GFP-TIP2;1 (parental control, A) or itt3 (B) were used for FRAP using a Zeiss710 confocal microscope and images were captured every 10 s. Images before (pre-bleach), immediately after (bleach 0’) and 6 min after (post-bleach 30’) bleaching are shown for one experiment. Bleached areas are shown with white (dashed) rectangles. ROIs that were used to measure fluorescence recovery are shown with colored rectangles. ROI fluorescence was quantified for complete vacuoles included in the bleach area (1, dark blue), vacuoles partially included in the bleach area (2, orange), non-bleached controls (3, light blue), and an area adjacent to the bleach area in partially bleached vacuoles (4, magenta). To measure the fluorescence recovery of vacuoles that were completely bleached, only the membrane adjacent to the cell wall was selected for quantification (1, dark blue). Bleaching was accomplished with an argon laser in the Zeiss LSM 710 microscope with excitation wavelength of 488 nm. The laser was used at 100% power and the pixel dwell time was 100.85 μsec. Scale bar: 20 μm. (C–D) Quantification of fluorescence recovery over time for GFP-TIP2;1 (parental control, C) or itt3 (D). Using 36 sets of FRAP experiments for each genotype as shown in A–C, the percent fluorescence recovery was calculated for each region of interest. The numbers and colors in the graphs correspond to those in A–B. N: 7 seedlings. Error bars indicate standard error.
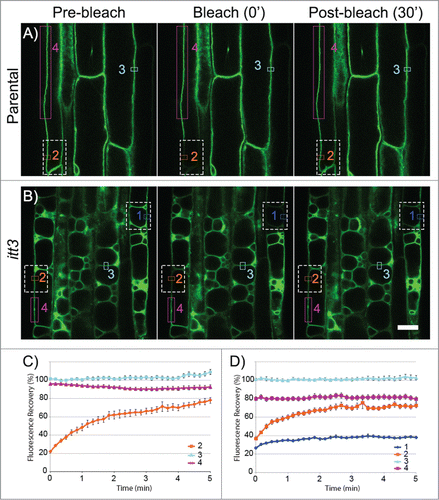
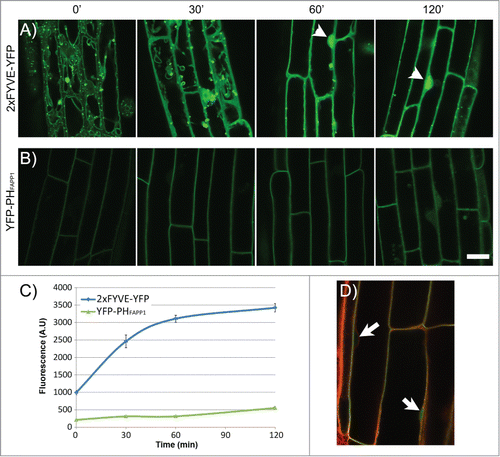
Figure 2. Effect of Wortmannin on the localization of YFP-2xFYVE and YFP-PHFAPP1 in the Arabidopsis root. (A–B) Time-lapse imaging of plant roots expressing YFP-2xFYVE (A) or YFP-PHFAPP1 (B) by confocal microscopy after Wm treatment. Seedlings were incubated for 0–120 min with 33 μM Wm. Acquisition settings were kept constant throughout the experiment in order to compare protein abundance between different time points. Bright nuclear signal is indicated with arrowheads. All images were captured on a Zeiss LSM710 confocal microscope. Scale bar: 20 μm. (C–D) Quantification of fluorescence signal in the nucleus in the 2xFYVE-YFP and YFP-PHFAPP1 during Wm treatment. Seedlings were treated as in (A) and stained with Lysotracker Red for 30 min before imaging. Nuclear signal for each marker was quantified using Zen software (Zeiss). N: 20 cells, 3 seedlings per data point. Bars represent standard error. (D) Lysotracker Red staining of the YFP-PHFAPP1 line to locate the nuclei (arrows).