Figures & data
Figure 1 Localization of promoter-GUS gene fusions in transgenic A. thaliana plants. (a) Seedling representative of PHT4;1-GUS and PHT4;4-GUS plants. (b) PHT4;2-GUS seedling. (c) Flower representative of PHT4;1-GUS, PHT4;4-GUS and PHT4;5-GUS plants. (d) PHT4;6-GUS flower. (e) Leaf and cotyledon representative of PHT4;3-GUS and PHT4;5-GUS plants showing vascular-specific GUS activity. (f) PHT4;6-GUS seedling. (g) Root of PHT4;1-GUS seedling. (h) Root of PHT4;3-GUS seedling. (i) Transverse leaf section representative of PHT4;1-GUS and PHT4;4-GUS plants. (j) Leaf epidermal peel representative of PHT4;1-GUS and PHT4;4-GUS plants. (k) Chlorophyll autofluorescence in epidermal peel shown in (j). (l) Transverse leaf section representative of PHT4;3-GUS and PHT4;5-GUS plants showing GUS activity in phloem. Bars in (g, h) = 100 µm; bars in (i-l) = 20 µm.
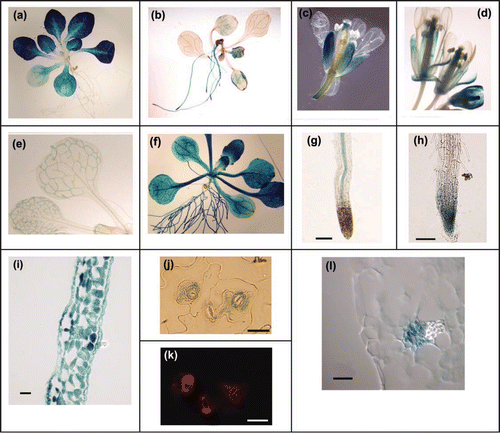
Figure 2 Effect of light on expression of PHT4 genes. Mature plants were held in the dark for 3 d before re-exposure to light. Expression levels were determined at the indicated time points by real-time RT-PCR and normalized to EIF-4A2. For each gene, expression values are relative to the maximum value, which was set at 1. The values plotted are averages of two biological replicates, and error bars indicate the replicate values. For analysis of PHT4;2, RNA was isolated from root tissues. For all other genes, RNA was isolated from rosette leaves. PHT2;1 serves as a positive control for light induction.
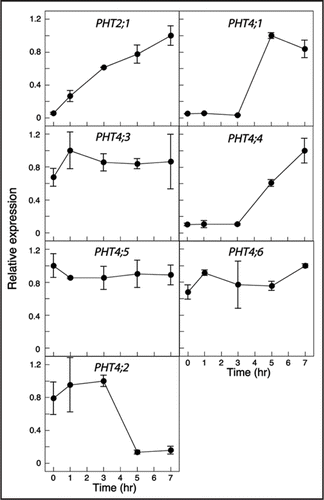
Figure 3 Expression of PHT4 genes under light/dark and constant light conditions. Expression levels were measured at the indicated times by real-time RT-PCR and normalized to EIF-4A2. For each gene, expression values are relative to the maximum value, which was set at 1. The values plotted are averages of two biological replicates, and error bars indicate the replicate values. The light and dark bars at the top of the figure indicate the respective light conditions. For analysis of PHT4;2, RNA was isolated from root tissues. For all other genes, RNA was isolated from rosette leaves.
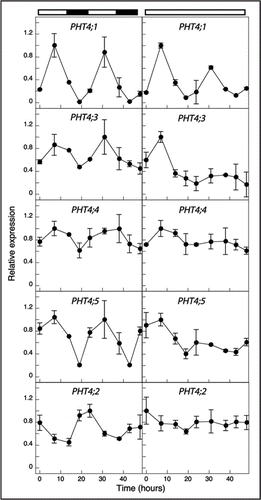
Figure 4 Neighbor joining tree of Arabidopsis and rice PHT4 Pi transporter proteins. Rice sequences are indicated by TIGR locus assignments. The numbers at the branches of the tree are bootstrap values (1,000 replicates). Scale bar indicates substitutions per site.

Table 1 Members of the rice PHT4 family