Figures & data
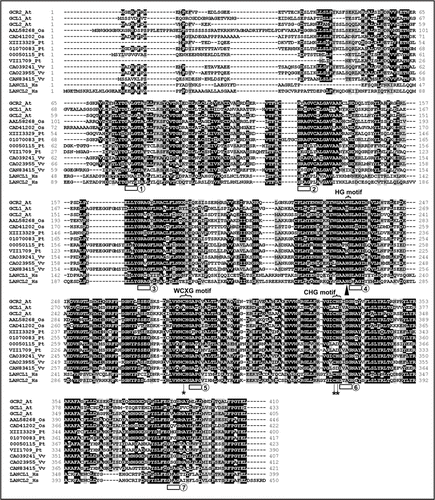
Figure 1 Amino acid sequence of GCR2 and multiple alignments with other lanthionine synthetase component C (LanC)-like proteins in plants and in humans. The amino acid sequences were aligned by CLUSTALW. Amino acids that are identical or similar are shaded with black or gray, respectively. The seven GXXG motifs are indicated by blocks underneath, and are numbered 1 to 7. The H reside of the HG motif in repeat 4 that is critical for substrate de-protonation for correct cyclization as shown in the crystal structure of the Lactococcus lactis LanC protein, nisin cyclise (NisC), and by site-directed mutagenesis studies,Citation18,Citation19 is indicated by a triangle underneath. The C residue of the WCXG motif of repeat 5 and the C and H residues in the CHG motif of repeat 6 that comprise conserved zinc-coordinating resides and are essential for the enzymatic activity of NisC, are indicated by stars underneath. The proteins aligned are (name of species and accession number in parentheses): GCR2_At (Arabidopsis thaliana, NP_175700), GCL1_At (Arabidopsis thaliana, NP_201331), GCL2_At (Arabidopsis thaliana, NP_850003), AAL58268_Os (Oryza sativa, AAL58268), CAD41202_Os (Oryza sativa, CAD41202), XIII3329_Pt (Populus trichocarpa, estExt_Genewise1_v1.C_LG_XIII3329), 01070083_Pt (Populus trichocarpa, eugene3.01070083), 00050115_Pt (Populus trichocarpa, eugene3.00050115), VIII1709_Pt (Populus trichocarpa, estExt_Genewise1_v1.C_LG_VII1709), CAO39241_Vv (Vitis vinifera, CAO39241), CAO23955_Vv (Vitis vinifera, CAO23955), CAN83415_Vv (Vitis vinifera, CAN83415), LANCL1_Hs (Homo sapiens, NP_006046) and LANCL2_Hs (Homo sapiens, NP_061167).