Figures & data
Figure 1 A model of the feedback regulation loop mediated by EIN3 and EBF2 in ethylene signaling. EIN3 is ubiquitinated by the SCFEBF complex and then degraded by the 26S proteasome. Ethylene represses this degradation pathway and the resulting accumulation of EIN3 activates the transcription of target genes. Simultaneously, EIN3 binds and activates the EBF2 promoter. EBF2 is recruited into an SCF complex (SCFEBF2) that promotes the ubiquitination and 26S proteasome-mediated degradation of EIN3.
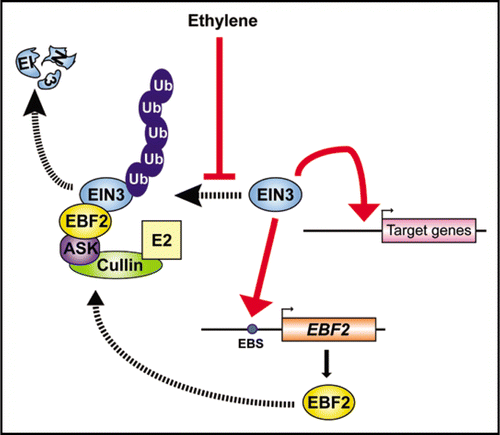
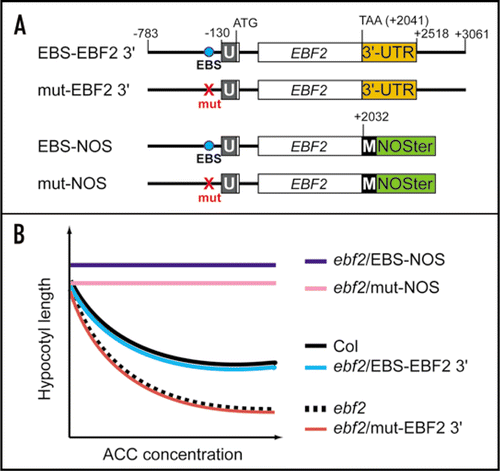
Figure 2 Requirement of two different regions of EBF2 for its precise expression. (A) Schematic representation of constructs used to transform the ebf2 mutant. These constructs contained either a DNA fragment corresponding to the region from −783 to +3061 of the EBF2 gene (relative to the translation initiation Met codon) or from −783 to +2032 fused to the NOS terminator (NOSter). The EIN3-binding site (EBS) and its mutated version are indicated by “EBS” and “mut”, respectively. The 5′ untranslated region and the MYC-tag are indicated by U and M, respectively. (B) Schematic representation of the ethylene sensitivity profiles of wild-type Columbia (Col), ebf2 and ebf2 transformed with the constructs listed in (A). Because ethylene induces a triple response that includes a shortening of the hypocotyls, the ethylene response is evaluated by measuring the hypocotyl lengths of etiolated seedlings that were grown in the presence of increased concentrations of 1-amino-1-cyclopropanecarboxylic acid (ACC), an immediate precursor of ethylene. Mutation of the EBS abolishes the complementation of the ethylene hyper-responsive phenotype of ebf2 (ebf2/mut EBF2 3′). The use of NOSter resulted in an ethylene-insensitive phenotype (ebf2/EBS-NOS and ebf2/mut-NOS). Note that the insertion of a MYC-tag at the 3′ end of the EBF2 ORF did not affect the ethylene sensitivity.Citation7