Figures & data
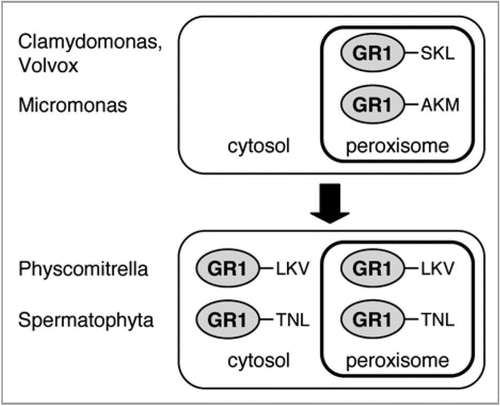
Figure 1 Analysis of PTS1 conservation in plant GR1 homologs. Sequences of full-length protein (FLP) plant GR1 homologs or ESTs (“EST”) were identified by BLAST and phylogenetic analysis, aligned by ClustalX, and conserved residues were shaded by Genedoc. In addition to spermatophyta, homologs from bryophyta and chlorophyta were analyzed for PTS1 conservation. For a phylogenetic analysis of the full-length proteins, see also Supplementary . The species abbreviations are as follows: Aa, Artemisia annua; At, Arabidopsis thaliana; Bn, Brassica napus; Br, Brassica rapa; Ci, Cichorium intybus; Cr, Chlamydomonas reinhardtii; Cs, Cynara scolymus; Fv, Fragaria vesca; Ha, Helianthus annuus; Msp, Micromonas sp. RCC 299; Mt, Medicago truncatula; Nt, Nicotiana tabacum; Os, Oryza sativa; Pk, Picrorhiza kurrooa; Ppat, Physcomitrella patens subsp. patens; Ps, Pisum sativum; Ptri, Populus trichocarpa; Rc, Ricinus communis; Rs, Raphanus sativus; Tp, Trifolium pratense; Tpus, Triphysaria pusilla; Vc, Volvox carteri f. nagariensis; Vv, Vitis vinifera; Zm, Zea mays.
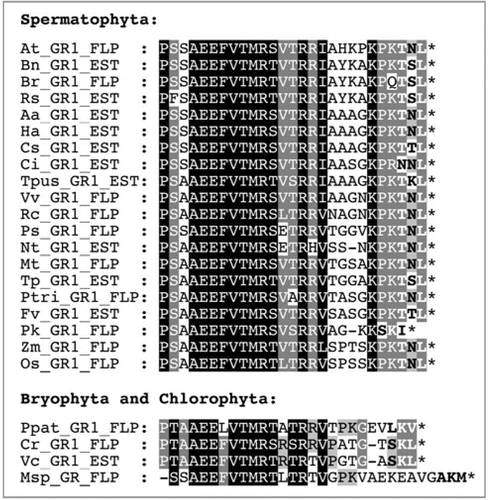
Figure 2 In vivo subcellular targeting analysis of Arabidopsis GR1. The full-length cDNA of Arabidopsis GR1 was fused N-terminally with the reporter protein EYFP and expressed transiently in onion epidermal cells upon biolistic bombardment or in tobacco protoplasts upon polyethylene glycol-mediated transformation. To characterize TN L> as the PTS1 of GR1, EYFP was extended C-terminally by the predicted peroxisome targeting domain of GR1, comprising the C-terminal 10 aa residues (AHKPKPKTNL>). In double transformants, peroxisomes were labeled with gMD H-CFP,Citation28 and the cyan fluorescence was converted to red to facilitate the detection of green and cyan fluorescent peroxisomes as yellow organelles in image overlays (merge). EYFP alone and EYFP extended C-terminally by 10 glycine residues (EYFP-10G) served as negative controls to verify the absence of endogenous bacteria. Scale bar: 10 µm.
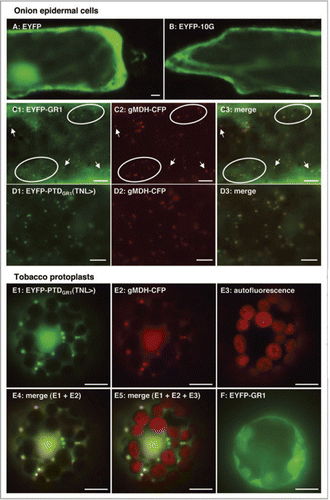
Figure 3 Model of the evolutionary development of the peroxisomal and cytosolic functions of GR1 in spermatophyta. The canonical nature and high peroxisome targeting strength of PTS1s in GR homologs of chlorophyta (Chlamydomonas, Volvox and Micromonas) indicate an exclusive compartmentalization of GR1 in peroxisomes in green algae. By contrast, the non-canonical nature and predicted (LKV>) and proven (TN L>) low peroxisome targeting strength in Physcomitrella and higher plants, respectively, indicate dual enzyme targeting to peroxisomes and the cytosol in bryophyta and spermatophyta.