Figures & data
Figure 1 Alignment of HSP70 proteins from selected crop species: Protein sequences were analyzed using DNA analysis software DNASTAR. For protein alignment, Clustal V of MegAlign was used in which clusters were aligned as pairs, then collectively as sequence groups to produce the overall alignment. Comparison of PgHsp70 with deduced amino acid sequences of chloroplast of Oryza sativa (accession no. ABA97211), Pisum sativum (accession no. CAA 49147), Cucumix sativus (accession no. ABM92419) and Arabidopsis thaliana (accession no. Np194159).

Figure 2 Sequence alignment of PgHsp70 and protein sequence corresponding to the template (1YUW). The domain regions such as nucleotide binding, linker and substrate binding are boxed and marked. Stars and dots in the sequences indicate the positions of identical amino acids and semi conserved substitutions, respectively.
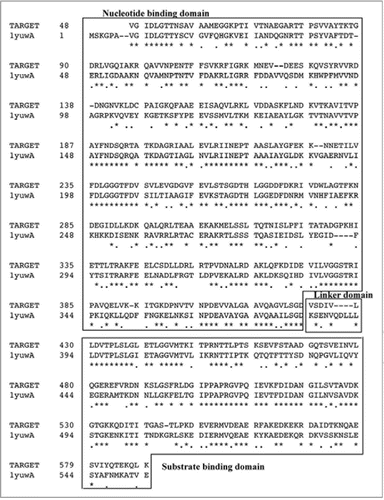
Figure 3 (A and B) Ribbon representation of PgHsp70 comparative model, Helix A highlighted in a box. The region above Helix A corresponds to the substrate binding domain (SBD) and the region below, to the nucleotide binding domain (NBD). Interdomain linker represented as red Cα trace. (C) Detailed view of interactions between SBD (Cα trace in red orange) and NBD (Cα trace in Cyan) and few discontinuous residues without Cα trace, side chains of residues identical or conserved to the template in blue, red, yellow or dark grey for +, −, polar, or nonpolar. PgHsp70 residues not conserved with respect to the template residues are shown in green.
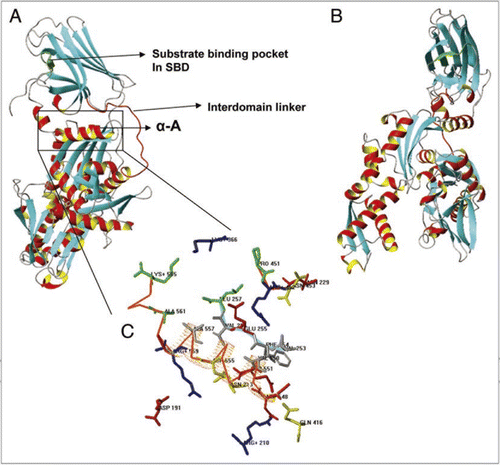
Figure 4 (A) SDS-PAGE analysis of purified recombinant PgHsp70 protein. PgHsp70 carrying a N-terminal hexahistidine tag was overproduced in pET-28a(+) and purified by Ni2+ NTA affinity chromatography. Lane 1 is molecular mass markers; the sizes (in kDa) are indicated adjacent (left side) to the gel. Lane 2 IPTG-induced supernatant fraction containing enriched PgHsp70 protein (70 kDa). Lane 3 is un-induced protein PgHsp70, Lane 4 is Ni2+ NTA purified PgHsp70. The gel containing lanes 1–4 is stained with Coomassie Brilliant Blue. Lane 5: the purified PgHsp70 protein was run on another gel and silver stained. (B) Reconfirmation of PgHsp70 purity by the reverse-phase-HPLC analysis (detector: 280 nm) on C18 (Phenomenex, C18, 5 μM 1.D. 250 × 4.6 nm) using aceto-nitrile (0.1% TFA)/water (0.1% TFA) gradient 0−15 min at the rate of 2% min−1 solvent; 15 to 45 min 0.5% min−1 solvent at a flow rate of 1.0 ml min−1.
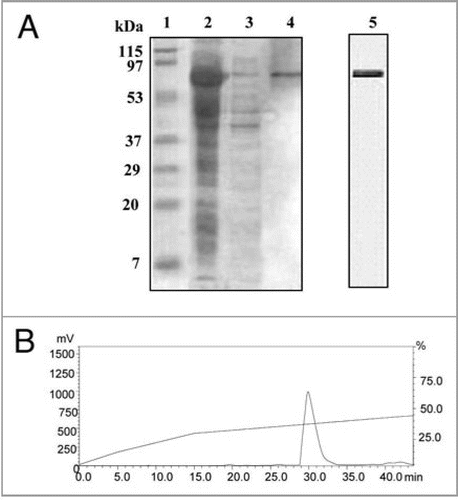
Figure 5 Thermally induced aggregation of model proteins such as glutamate dehydrogenase/or alcohol dehydrogenase with/without PgHsp70 at 50°C was Light scatter assayed by incubating the reaction mixture (1 ml) containing 20 mM Tris buffer (pH 8.0), 100 mM sodium chloride, 0.2 mg ml−1 glutamate dehydrogenase/or alcohol dehydrogenase and 0.2 mg ml−1 of the recombinant heat shock protein PgHsp70 (with or without ATP 5 mM) or BSA (0.2 mg ml−1) aggregation was monitored by the measuring the O.D. at 360 nm. (A) Light scatter assay was done using substrate protein GDH to assess the chaperone activity of PgHsp70 in the presence and absence of BSA (B) Light scatter assay was done using substrate protein ADH to assess the chaperone activity of PgHsp70 in the presence and absence of BSA. (C) Light scatter assay was done using substrate protein GDH to assess the chaperone activity of PgHsp70 in the presence and absence of ATP. The error bars have not been added in this figure because the SD in all the cases was less than 10%.
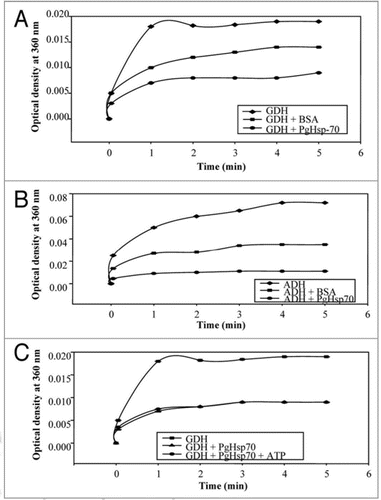
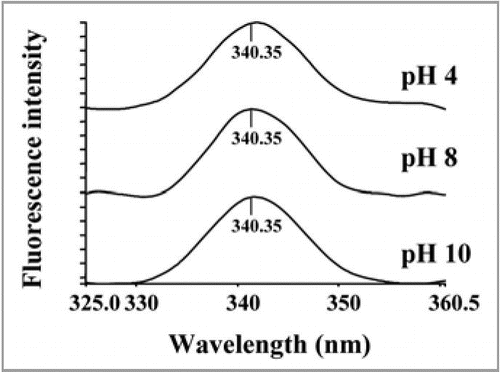
Figure 6 Effect of pH (A) on chaperone activity of PgHsp70; (B) on stability of PgHsp70, (C) effect of temperature on chaperone activity (experimental conditions as mentioned in ), and (D) temperature stability of PgHsp70.
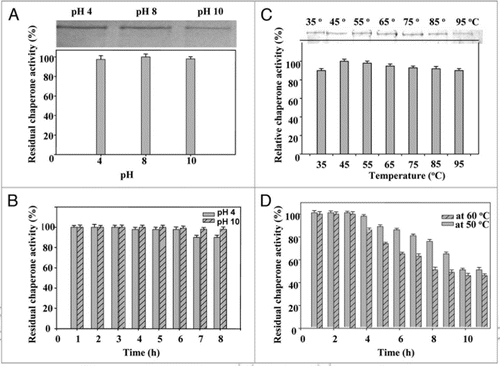
Figure 7 Effect of anion charger KI: (A) on chaperone activity of native PgHsp70 in 0.1 M phosphate buffer (pH 8.0) at 50°C for 5 min. (B) on secondary structure of PgHsp70 [spectra 1 & 2–5 represents native PgHsp70, different concentration of KI (0.2–0.5 mM) with native PgHsp70] (C) Effect of acrylamide on chaperone activity experimental conditions as mention in . The error bars have not been added in this figure because the SD in all the cases was less than 10%.
![Figure 7 Effect of anion charger KI: (A) on chaperone activity of native PgHsp70 in 0.1 M phosphate buffer (pH 8.0) at 50°C for 5 min. (B) on secondary structure of PgHsp70 [spectra 1 & 2–5 represents native PgHsp70, different concentration of KI (0.2–0.5 mM) with native PgHsp70] (C) Effect of acrylamide on chaperone activity experimental conditions as mention in Figure 5. The error bars have not been added in this figure because the SD in all the cases was less than 10%.](/cms/asset/d8ceca4b-412e-40da-a64c-e1314ae1c6ef/kpsb_a_10910547_f0007.gif)
Figure 8 Effect of inhibitor Gdn.HC l: (A) on secondary structure of PgHsp70 [spectra 1–5 represent PgHsp70, PgHsp70 with different concentrations of Gdn.HC l (0.2 to 0.5 mM)], (B) on chaperone activity of PgHsp70 (experimental conditions as mentioned in ).
![Figure 8 Effect of inhibitor Gdn.HC l: (A) on secondary structure of PgHsp70 [spectra 1–5 represent PgHsp70, PgHsp70 with different concentrations of Gdn.HC l (0.2 to 0.5 mM)], (B) on chaperone activity of PgHsp70 (experimental conditions as mentioned in Fig. 5).](/cms/asset/fdfeb755-e9eb-48cb-b394-dc18425b9cf3/kpsb_a_10910547_f0008.gif)
Figure 9 Effect of urea: (A) on secondary structure of PgHsp70 spectra 1–3 represents different concentrations of urea (0.2 to 0.4 mM) with PgHsp70 (B) on Chaperone activity of native PgHsp70 (experimental conditions as mentioned in ).
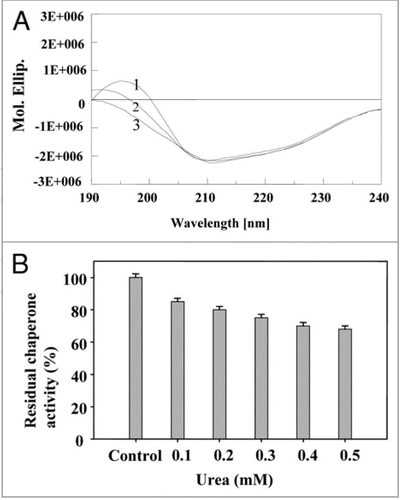
Figure 10 Effect of SDS: (A) on secondary structure of PgHsp70 [spectra 1–5 represents PgHsp70 and different concentration of SDS (0.2 to 0.5%) with PgHsp70 (B) on chaperone activity (experimental conditions as mentioned in ).
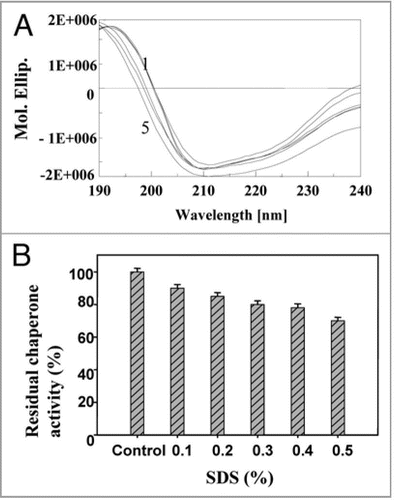
Figure 12 Coomassie brilliant blue-stained SDS-PAGE gel. Electrophoretic fractionation of simultaneous overexpression of PgHsp70 along with CA prevents the aggregation and accordingly prevents the formation of inclusion bodies in E. coli: Lane 1 is molecular weight of marker lane; the sizes (in kDa) are indicated adjacent (left side) to the gel. Lane 2 is co-expression of CA with PgHsp70 showing the high level of overproduction of CA in supernatant after after 3 h induction with IPTG (IPTG induced supernatant fraction). Lane 3 is induced CA in the pellet fraction (without PgHsp70); i.e., without co-expression the CA protein was going to pellet fraction. Lane 4 is a pellet fraction of only PgHsp70. Lane 5 is pellet fraction after co-expression of CA with PgHsp70; the CA and PgHsp70 proteins are not appearing. Lanes 4 and 5 were run in another gel and Coomassie Brilliant Blue stained.
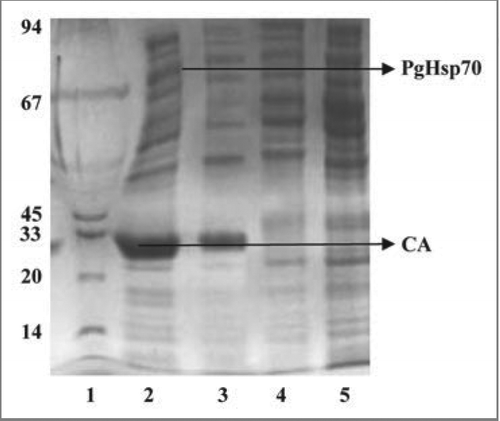
Table 1 The relative fold change of PgHsp70 transcript under different abiotic stress conditions
Table 2 Effect of different physical (temperature and pH) and chemical agents (denaturants and inhibitors) on secondary structure of PgHsp70