Figures & data
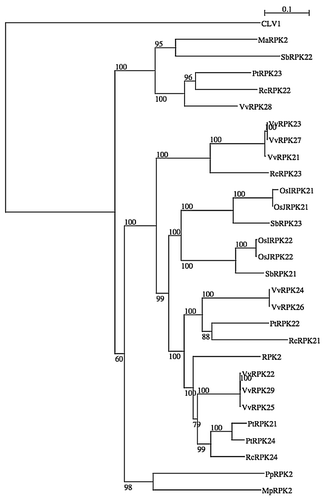
Figure 1 A neighbor-joining tree of RPK2. Protein sequence of RPK2 homologues of Arabidopsis thaliana (RPK2 and CLV1 as an out group), Musa acuminata (MaRPK2: ABF72006.1), Oryza sativa Indica Group (OsIRPK21: EAY91907.1, OsIRPK22: EAZ04623.1), Oryza sativa Japonica Group (OsJRPK21: NP_001051315.1, OsJRPK22: NP_001060207.1), Populus trichocarpa (PtRPK21: XP_002305358.1, PtRPK22: XP_002311344.1, PtRPK23: XP_002321080.1, PtRPK24: XP_002323902.1), Vitis vinifera (VvRPK21: CAN67126.1, VvRPK22: CAN77668.1, VvRPK23: CAO40245.1, VvRPK24: CAO41857.1, VvRPK25: CAO49170.1, VvRPK26: XP_002274047.1, VvRPK27: XP_002274211.1, VvRPK28: XP_002276030.1, VvRPK29: XP_002279979.1), Sorghum bicolor (SbRPK21: XP_002460974.1, SbRPK22: XP_002462751.1, SbRPK23 XP_002463860.1), Ricinus communis (RcRPK21: XP_002512071.1, RcRPK22: XP_002512822.1, RcRPK23: XP_002515143.1, RcRPK24: XP_002527617.1), Physcomitrella patens (PpRPK2: XP_001781758.1) and Marchantia polymorpha (MpRPK2: isotig22424), were used to construct the phylogenic tree. Bootstrap values of 60% and above, from the neighbor-joining method with Kimura's correction, are shown. The scale bar indicates the number of amino acid substitutions per site.