Figures & data
Figure 1. Splicing of exons 5a to 5b is induced by an IE1 mutation, but not an IE1 deletion in B. mori 14–3-3ζ pre-mRNA. (A) Genomic organization of the B. mori (Bmo) 14–3-3ζ gene. Constitutive exons (in black boxes), alternative exons (in blue boxes), conserved intronic elements IE1 and IE2 (yellow), and the intron (line) are shown. Docking sites (marked by saddle shapes) are reverse-complementary to an upstream selector sequence (marked by hearts). (B) Effects of mutations on exon 5a and exon 5b inclusion. (C) Effects of mutations on exon 5a and exon 5b selection. The specific RT-PCR band containing exon 5a or exon 5b was purified for digestion using an exon-specific restriction enzyme. (D). Schematic diagrams of the mutant constructs that test for splicing. A series of constructs were made, with IE1 replaced by inserts of 6, 13, 18, 24, 31, and 35 nt. (E). Effects of inserts on exon 5 splicing. (F). Schematic diagrams of the minigene constructs used to test the importance of the RNA hairpin by increasing the loop sizes. (G). Effect of the enlarged loops on exon 5 selection. Data are expressed as the percentage of the mean ± standard deviation from three independent experiments.
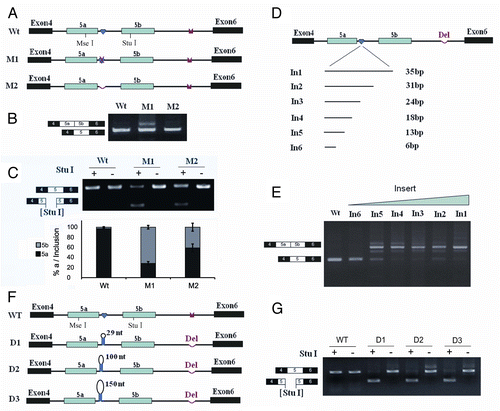
Figure 2. Hairpins are evolutionarily conserved in lepidopteran species. Sequences aligned from the indicated lepidopteran species (for abbreviations used, see Supp Materials). Symbols used are the same as in . Identical nucleotides at each position are shown in different colors. RNA hairpins within IE1 and the intronic RNA pairings between the selector sequences and the docking site were predicted using the Mfold program.Citation25
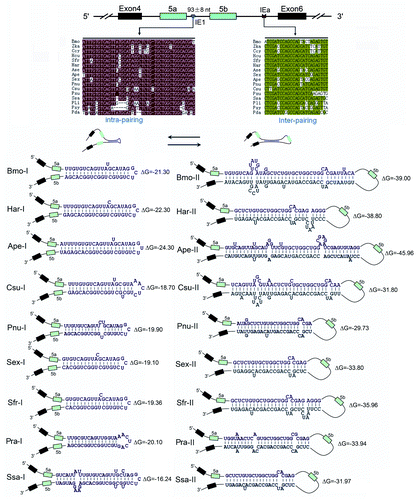
Figure 3. Regulation of mutually exclusive splicing by competitive RNA pairing between intra- and inter-introns. (A) Genomic organization of the 14–3-3ζ gene of silkworm. The symbols used are the same as in . (B) Schematic diagrams of mutant constructs for splicing. The predicted mutually exclusive RNA pairings are shown. Mutations introduced into dsRNA are indicated (M1-M6). Also shown are the predicted RNA pairings for wild-type RNA and a series of mutants (point mutations are shown in boldface). The arrow depicts the suppression of exon 5b. (C) Modulation of exon 5a selection by mutation analysis. (D) Correlation analysis of the RNA pairing competition between intra- and inter-introns using exon 5a selection.
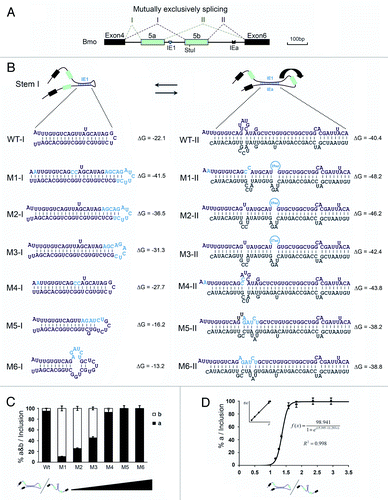
Figure 4. Correlation of the dynamic competition between intra- and inter-intronic RNA pairing with the ratio of exon 5a/5b. Correlation analysis of intra-intronic RNA pairing strength (A), inter-intronic RNA pairing strength (B), and their relative strength (C) with the ratio of exon 5a in Lepidopteran species.
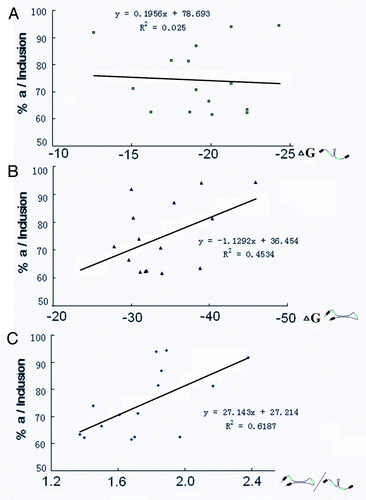
Figure 5. Effects of mutations and deletions on exon 5a inclusion. (A) Overview of the minigene constructs used to test the importance of inter-intronic RNA pairing. The red arrow in IE or IEa denotes the disrupting mutation. IE1, IE1, and IEa elements are indicated as in . WT: wildtype. (B) Effects of exon 5 selection by the series of deletions and mutations. Exon 5a is exclusively included in all constructs even though these constructs lack IE1 and IEa.
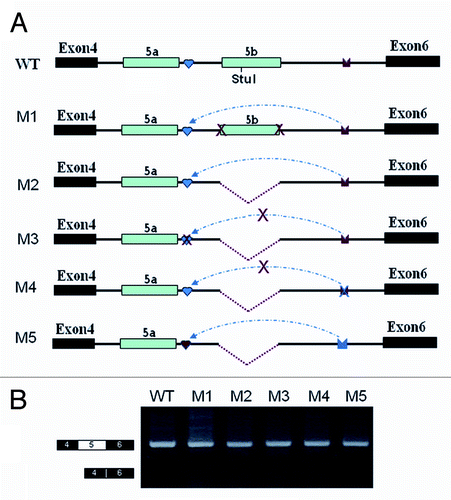
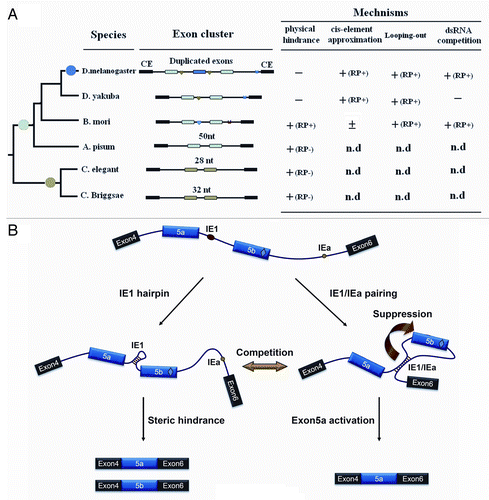
Figure 6. A refined model for regulating mutually exclusive splicing of silkworm 14–3-3ζ pre-mRNA. (A) Divergence of control mechanisms of mutually exclusive splicing. Exons that are duplicated in tandem are shown in colored boxes while constitutive exons (CE) flanking the duplicated exons are shown in black. Docking sites (marked by saddle shapes) are reverse-complementary to an upstream selector sequence (marked by hearts). The exon duplications (solid circles) produce the new exon copy. The introns are represented by lines, and not drawn to scale. “+” and “-” indicate the presence or absence, respectively, of underlying mechanisms for mutually exclusive splicing. “(RP+)” and “(RP-)” denote the RNA pairing-dependent and RNA pairing-independent mechanisms, respectively. “n.d” means “not determined.” (B) A refined model for controlling mutually exclusive splicing. The red and green circles represent the selector sequences and the docking site, respectively. The red diamond in the exon depicts the exonic enhancer. The arrow depicts the suppression of exon 5b. When the selector sequence (IE1) can interact with the docking site (IEa), the exon 5a variant is included in the 14–3-3ζ mRNA. Then inter-intronic RNA pairing excludes in-loop exon 5b via a looping-out mechanism. Alternatively, the RNA hairpin is assumed to form within the IE1 sequence. Thus, the resulting steric hindrance mechanism should dominantly control mutually exclusive splicing of these pre-mRNAs.