Figures & data
Figure 1. Organization of the pAD1 par locus and the Fst toxin. Converging promoters (black arrowheads labeled P) transcribe the toxin-encoding RNA I (red shaded arrow below line) and the antitoxin RNA II (dark green arrow above line) toward a bi-directional intrinsic transcriptional terminator (converging green arrows). The RNAs are transcribed across direct repeats DRa (pink arrows) and DRb (blue arrows) at which interaction occurs, suppressing translation of the FstpAD1 coding sequence (dark red box on DNA and RNA). The amino acid sequence of the Fst toxin is shown using standard single letter amino acid designations. The essential, conserved hydrophobic domain is shown in bold red print. This forms part of a transmembrane domain in the recently published structure of Fst.Citation22 The two blue underlined amino acids at the N-terminus must be charged to retain toxin function. The non-essential C-terminal tail is shown in green italics.
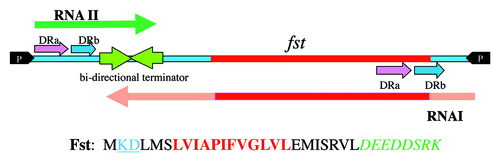
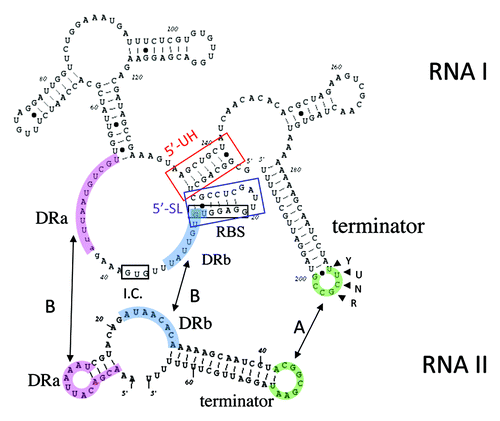
Figure 2. Secondary structures of RNA IpAD1 and RNA IIpAD1. The specific regions of interaction between the RNAs are shaded different colors to coordinate with and labeled accordingly. Interaction is initiated at the U-turn motif (labeled YUNR) present in the loop of the terminator of RNA I (green shaded). This interaction is indicated by the arrow labeled A. The interaction then extends to the direct repeat sequences DRa (pink shaded) and DRb (blue shaded). This interaction is indicated by arrows labeled B and is responsible for preventing translation of FstpAD1, since the initiation codon (I.C.) and the ribosome binding site (SD) are sequestered by the interacting RNAs. The two structures, 5′-SL (blue box) and 5′-UH (red box) are responsible for preventing premature translation of FstpAD1 and RNA IpAD1 stability, respectively.