Figures & data
Figure 1. The DEAD-box helicase Mss116p and its conserved sequence motifs. Upper panel: Schematic representation of the helicase core region of the DEAD-box protein Mss116p. Mt, mitochondrial localization signal, which is cleaved off after import; NTE – N-terminal extension; CTE, C-terminal extension; C-tail, containing numerous basic amino acids. Functional regions shown in black or white, respectively, are not part of the 3D structure. Lower panel: Crystal structure of the Mss116p helicase core, which consists of two RecA-like domains, and its helical C-terminal extension. The conserved motifs are colored according to their primary function: red, ATP binding and hydrolysis; blue, RNA binding; yellow, communication between ATP binding and RNA binding sites. The non-conserved regions of the helicase domains are in gray and the CTE is shown in light-gray. The RNA is shown in pale yellow, the non-hydrolyzable ATP analog (AMPNP) in white and the Mg2+ ion in green. This figure has been adapted from references Citation68,Citation72,Citation80. Note: in some recent helicase reviews on DEAD-box helicases, motif Ib has been renamed to motif Ic, while the GG doublet has become motif Ib, thus we also refer to the GG doublet as motif Ib and to the TPGRLID sequence as motif Ic as described in references Citation64,Citation66,Citation80.
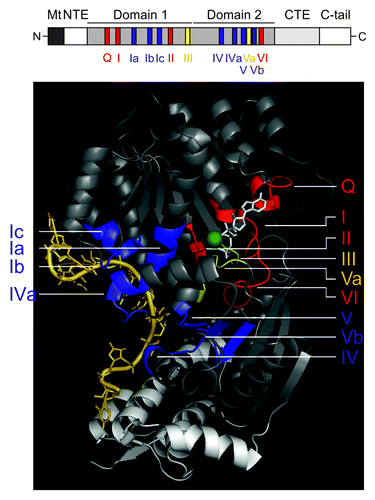
Table 1. Effects of mutations on Mss116p function
Figure 2. Interactions of the ATP analog and ssRNA with the helicase core. Left panel: Amino acids that were mutated to dissect Mss116p’s function in splicing of yeast mitochondrial introns are highlighted: K158, violet; S305, cyan; T307, dark cyan; Q412, blue; R245, red; I551, green; K569 – orange; the RNA is colored in pale yellow and AMPNP in white. The Mg2+ ion is represented as bright yellow sphere. Note that I551 and K569 are shown to indicate the start site of truncations in respective Mss116p mutants. Right panel: Conserved amino acids of the RecA-like domains contacting the non-hydrolyzable ATP analog (AMPNP; upper right panel) and RNA (lower right panel), respectively.
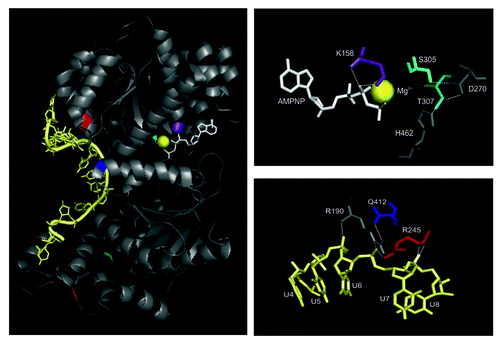
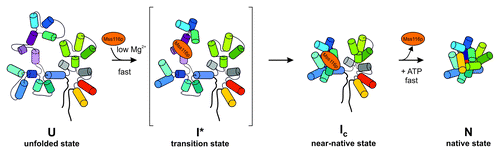
Figure 3. Mss116p-facilitated folding of the ai5γ intron in vitro. The DEAD-box protein accelerates compaction of intron domain D1 to a near-native folding intermediate, by stabilizing a specific structure at the core of this intron domain early in the ai5γ folding pathway. Subsequently, other intron domains can dock rapidly onto the D1 scaffold, whereby Mss116p does not stabilize the native state and is recycled upon ATP hydrolysis. D1 is shown in blue shades except for the κ−ζ element colored in purple shades, D3 is in green, D5 in red and D6 in yellow, while D2 and D4 are depicted in gray. Exons are outlined as thick black lines. Mss116p is indicated as orange ellipse. This figure has been adapted from references Citation52,Citation53.