Figures & data
Figure 1. Ibs-Sib loci in E. coli MG1655. (A) Genetic organization of the loci. (B) Alignment of the five sib gene sequences from E. coli MG1655. The Target Recognition Domains (TRD1 and TRD2) are indicated as determined by Han, et al.Citation16
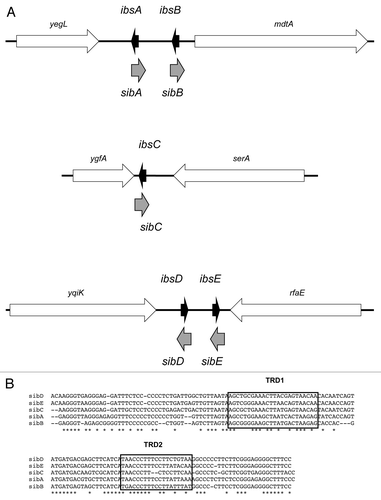
Figure 2. ShoB-OhsC locus of E. coli MG1655. (A) Genetic organization of the locus. Note the arrow for shoB indicates the mRNA; the box within the arrow indicates the coding region of the sequence. (B) Base pairing between shoB and OhsC. (C) Predicted secondary structure of shoB.Citation28 The start codon is boxed; the predicted ribosome binding site is indicated by the dashed bracket; the region of base pairing to OhsC is indicated by the bracket.
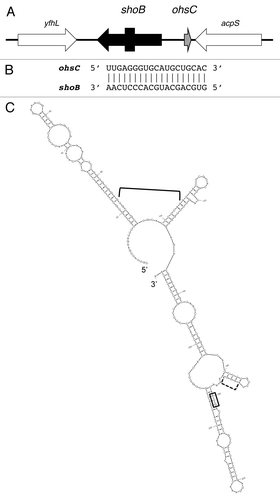
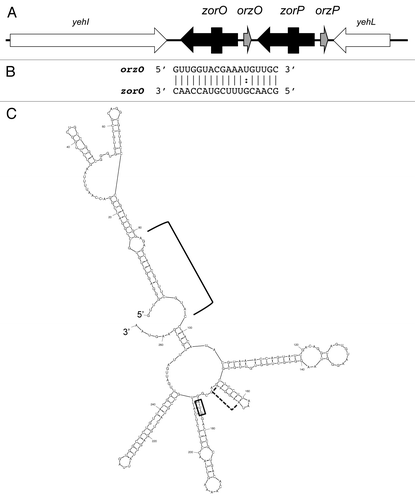
Figure 3. Zor-Orz locus of E. coli O157:H7 str. EDL933. (A) Genetic organization of the locus. Note the arrows for zorO and zorP indicate the mRNAs; the boxes within the arrows indicate the coding region of the sequences. (B) Base pairing between zorO and OrzO. (C) Predicted secondary structure of zorO.Citation28 The start codon is boxed; the predicted ribosome binding site is indicated by the dashed bracket; the region of base pairing to OhsC is indicated by the bracket.