Figures & data
Figure 1. Schematics of the tisB-istR1 locus with RNAs encoded. A. The locus is drawn schematically, with promoters (angled arrows), terminators (stemloops) and LexA box indicated. The three primary transcripts are shown. B. RNAs observed under SOS-ON and SOS-OFF (LexA-repressed) conditions. The three mRNA variants (blue) are indicated. “X” at the 5′-end indicates a processing step. In SOS-ON, also IstR1 is processed. See text for details.
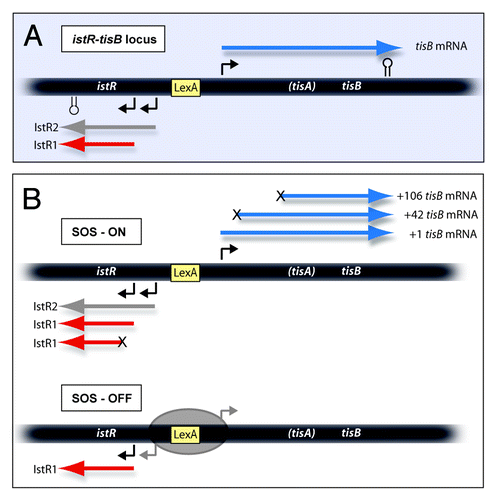
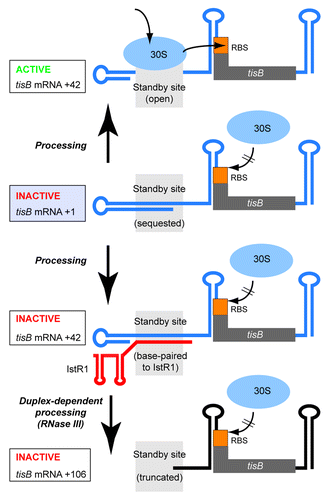
Figure 2. Standby requirement for tisB translation and inhibition by IstR1. Inactive +1 tisB mRNA (light blue box) is processed into +42 (up and down arrow). Arrow up: An open standby region (gray field) permits binding of standby 30S subunits, relocation and translation initation at the tisB RBS. Arrows down: Binding of IstR1 blocks the standby site, subsequent RNase III cleavage removes part of it. See text for details.