Figures & data
Figure 1. LeishIF4E-3 interacts with LeishIF4G-4. Yeast two-hybrid assays were performed by co-transfection of wild typeYRG2 cells with plasmids encoding the Gal-4 Activation Domain (AD) fused to LeishIF4G-1 to -6 (LmjF30.1150, LmjF15.1320, LmjF16.1600, LmjF36.6060, LmjF10.1080 and LmjF15.0060, denoted 4G1 to 6, respectively) and the Gal-4 Binding Domain (BD) fused to LeishIF4E-3 (denoted 4E3). The cells were cultured under permissive (+His) and restrictive (-His) conditions and spotted in 3-fold dilutions. Expression of exogenous proteins in the transfected yeast strains is shown in Figure S1.
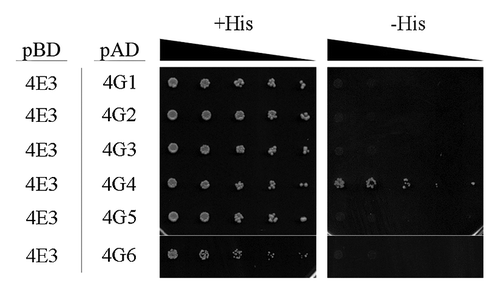
Figure 2. Reciprocal pull-down of LeishIF4E-3 and LeishIF4G-4. Transgenic L. amazonensis cells expressing SBP-tagged LeishIF4G-4 (A) or TAP-tagged LeishIF4E-3 (B) were grown under normal conditions and further subjected to nutritional starvation for 6 h. Following washes and cell disruption, protein complexes were affinity-purified over streptavidin-sepharose, separated by SDS-PAGE and immunoblotted with antibodies against LeishIF4G-4 or LeishIF4E-3. The lanes were loaded with 1% [supernatant (S), flow through (F)] or with 20% [wash (W), elution (E)] of the total experimental sample. (C) A list of proteins related to translation identified by mass spectrometry in the pulled-down complexes of tagged LeishIF4E-3 and LeishIF4G-4. The complexes were extracted from promastigotes grown under normal conditions. The results represent data from two independent experiments. The complete list of proteins is provided in Table S1.
![Figure 2. Reciprocal pull-down of LeishIF4E-3 and LeishIF4G-4. Transgenic L. amazonensis cells expressing SBP-tagged LeishIF4G-4 (A) or TAP-tagged LeishIF4E-3 (B) were grown under normal conditions and further subjected to nutritional starvation for 6 h. Following washes and cell disruption, protein complexes were affinity-purified over streptavidin-sepharose, separated by SDS-PAGE and immunoblotted with antibodies against LeishIF4G-4 or LeishIF4E-3. The lanes were loaded with 1% [supernatant (S), flow through (F)] or with 20% [wash (W), elution (E)] of the total experimental sample. (C) A list of proteins related to translation identified by mass spectrometry in the pulled-down complexes of tagged LeishIF4E-3 and LeishIF4G-4. The complexes were extracted from promastigotes grown under normal conditions. The results represent data from two independent experiments. The complete list of proteins is provided in Table S1.](/cms/asset/1379653f-fce0-4095-ae0e-174eaa1f272e/krnb_a_10922709_f0002.gif)
Figure 3. LeishIF4E-3 changes its pattern of expression only during nutritional stress. Wild type L. amazonensis promastigotes were subjected to nutritional starvation or heat shock (33°C) for 3, 6 or 12 h. Whole cell extracts were separated by SDS-PAGE and subjected to western analysis using specific antibodies against LeishIF4E-1, LeishIF4E-3 or LeishIF4G-4. LeishIF4E-1, which remains unchanged throughout differentiation, served as the loading control.
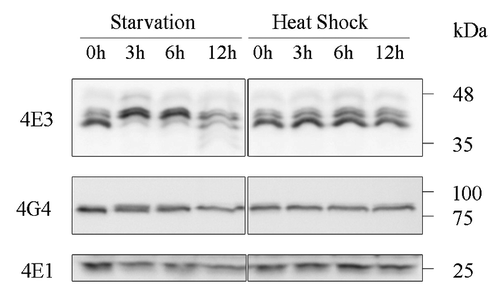
Figure 4. Differential expression of LeishIF4E-3 and LeishIFG-4 in promastigotes and axenic amastigotes. Whole cell extracts of wild type L. amazonensis late-log phase promastigotes (P) and axenic amastigotes, 9 d after differentiation was initiated (A), were separated by SDS-PAGE and subjected to western analysis using specific antibodies against GP46, HSP100, LeishIF4E-1, LeishIF4E-3 or LeishIF4G-4. LeishIF4E-1, which remains unchanged throughout differentiation, served as the loading control.
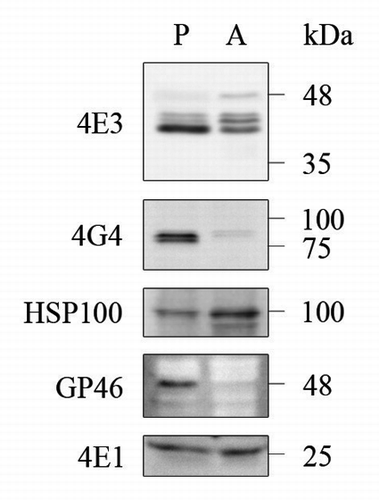
Figure 5. LeishIF4E-3 and LeishIF4G-4 migration in sucrose gradients. Extracts of wild type L. amazonensis cells were fractionated over 10–40% sucrose gradients. The OD260 of the different gradient fractions is shown in the upper panels. Proteins were immunoblotted with antibodies specific for LeishIF4E-3 or LeishIF4G-4. Samples were obtained from the supernatants (S) or from TCA-precipitated gradient fractions. The experiment was performed on promastigotes grown under normal conditions (A) or subjected to nutritional starvation for 6 h (B).
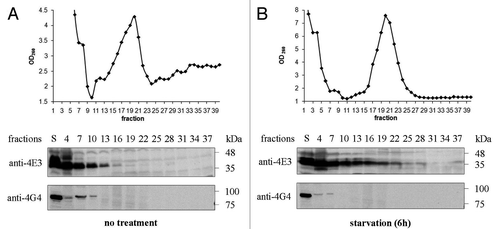
Figure 6. LeishIF4E-3 enters granules that do not contain LeishIF4G-4. Wild type L. amazonensis cells were subjected to heat shock or nutritional starvation for 3 and 6 h (presented in Fig. S6). The average number of granules per cell was counted and standard deviations were calculated for wild type cells experiencing heat shock (A) or nutrient starvation (B). LeishIF4G4-GFP-expressing L. amazonensis cells were subjected to heat shock or nutritional starvation for 3 and 6 h (C). LeishIF4E-3 was detected by a specific antibody and a secondary fluorescent antibody (DyLight). LeishIF4G-4 was visualized through its fusion with GFP. Nuclear and kinetoplast DNA were visualized by Hoechst staining. The average number of granules per cell was obtained as in A and B, for cells subjected to heat shock (D) or nutrient starvation (E).
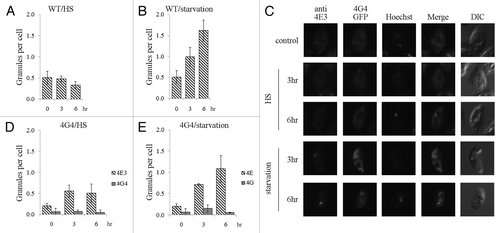
Figure 7. LeishIF4E-3 enters stress granules during starvation. L. amazonensis cells expressing LeishDHH1-GFP (A) were subjected to heat shock or nutritional starvation for 3 and 6 h. LeishIF4E-3 was detected by a specific antibody and a secondary fluorescent antibody (DyLight). LeishDHH1 was visualized through its GFP fusion. Nuclear and kinetoplast DNA were visualized by Hoechst staining. The average number of granules per cell was counted and standard deviations were calculated for LeishIF4G4-GFP-expressing cells subjected to heat shock (B) and nutrient starvation (C). A similar analysis was performed for wild type cells experiencing nutritional stress, with or without cycloheximide (CHX) (D).
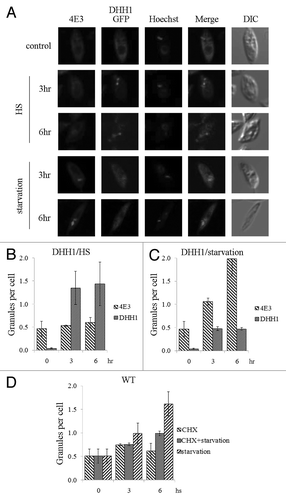
Figure 8. Binding to LeishIF4E-3 is mediated by a short non-conserved peptide in the N-terminus of LeishIF4G-4. LeishIF4G-4 regions that interact with LeishIF4E-3 were identified using a yeast two-hybrid assay. (A, B) Wild type YRG2 cells were co-transfected with the yeast plasmids encoding AD and BD fusion proteins. The BD sequence was fused to the open reading frame of LeishIF4E-3. The AD sequence was fused to (A) the complete open reading frame of LeishIF4G-4, LeishIF4G-4(1–78), LeishIF4G-4(79–335), LeishIF4G-4(336–765), LeishIF4G-4-N (1–78) or to (B) the N-terminus of LeishIF4G-4, denoted LeishIF4G-4-N(1–78), and to LeishIF4G-4-N containing deletions Δ2–16, Δ17–31, Δ32–46, Δ63–78, Δ17–21, Δ22–26 and Δ27–31. The ability (+) or inability (-) of the yeast strains to growth on restrictive medium are marked in the Interaction column. (C) The LeishIF4G-4 peptide fragment that was mapped to be essential for binding of LeishIF4E-3 is shown in bold. The yeast growth on plates is shown in Figure S2 and expression of the exogenous proteins in the transfected yeast strains is shown in Figure S1.
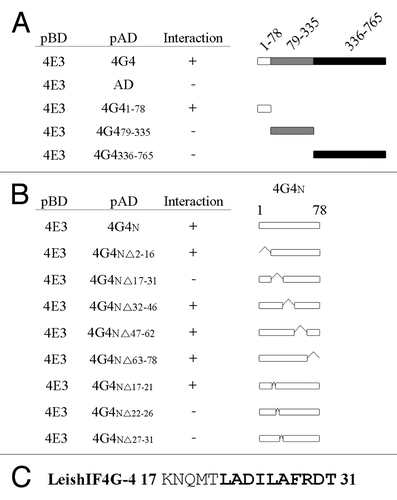
Figure 9. LeishIF4E-3 binding depends on the structure of the conserved peptide rather than on its sequence. (A) Multiple sequence alignment of the LeishIF4E-3 binding region in LeishIF4G-4 derived from different trypanosomatid species [L. major (LmjF36.6060), L. infantum (LinJ.36.6320), L. braziliensis (LbrM.35.6370), L. tarentolae (LtaP36.6220), T. brucei (Tb11.01.2330) and T. cruzi (Tc00.1047053510285.100)]. The amino acids of the fragment that was shown to be essential for binding are shown in bold. (B) Point mutation analysis of LeishIF4G-4 amino acids 22–31, using yeast two-hybrid assays. Wild type YRG2 cells were co-transfected with plasmids encoding the AD and BD fusion proteins. The AD sequence was fused to the complete open reading frame of LeishIF4G-4 and to its N-terminus LeishIF4G-4-N (1–78), as well as to LeishIF4G-4-N (1–78) carrying the following mutations: L22A, D24A, I25A, L26A, F28A, R29A, D30A and T31A. The BD sequence was fused to wild type LeishIF4E-3 and to the LeishIF4E-3(W187A) mutant. The ability (+) or inability (-) of the yeast strains to growth on restrictive medium are marked in the Interaction column. (C) Secondary structure prediction of the LeishIF4G-4 region found to be essential for binding of LeishIF4E-3, as generated by the PsiPred server. Alpha helices (H) are shown as purple cylinders, and the confidence of the prediction is shown above (higher and darker blue columns represent higher confidence). Predictions are shown for the wild type sequence, as well as for the L26A and L26P mutants. (D) Mutational analysis of L26 in LeishIF4G-4 using a yeast two-hybrid assay. The assays were performed as described in B. The AD sequence was fused to the LeishIF4G-4-N (1–78) fragment carrying the L26A and L26P mutations. The BD sequence was fused to LeishIF4E-3. The yeast growth on plates is shown in Figure S2 and expression of the exogenous proteins in the transfected yeast strains is shown in Figure S1. (E) Transgenic L. amazonensis cells expressing SBP-tagged LeishIF4G-4 or LeishIF4G-4(L26P) were grown under normal conditions. Following washes and cell disruption, protein complexes were affinity-purified over streptavidin-sepharose, separated by SDS-PAGE and immunoblotted with antibodies against LeishIF4G-4 or LeishIF4E-3. The lanes were loaded with 1% [supernatant (S), flow through (F)] or with 20% [wash (W), elution (E)] of the total experimental sample.
![Figure 9. LeishIF4E-3 binding depends on the structure of the conserved peptide rather than on its sequence. (A) Multiple sequence alignment of the LeishIF4E-3 binding region in LeishIF4G-4 derived from different trypanosomatid species [L. major (LmjF36.6060), L. infantum (LinJ.36.6320), L. braziliensis (LbrM.35.6370), L. tarentolae (LtaP36.6220), T. brucei (Tb11.01.2330) and T. cruzi (Tc00.1047053510285.100)]. The amino acids of the fragment that was shown to be essential for binding are shown in bold. (B) Point mutation analysis of LeishIF4G-4 amino acids 22–31, using yeast two-hybrid assays. Wild type YRG2 cells were co-transfected with plasmids encoding the AD and BD fusion proteins. The AD sequence was fused to the complete open reading frame of LeishIF4G-4 and to its N-terminus LeishIF4G-4-N (1–78), as well as to LeishIF4G-4-N (1–78) carrying the following mutations: L22A, D24A, I25A, L26A, F28A, R29A, D30A and T31A. The BD sequence was fused to wild type LeishIF4E-3 and to the LeishIF4E-3(W187A) mutant. The ability (+) or inability (-) of the yeast strains to growth on restrictive medium are marked in the Interaction column. (C) Secondary structure prediction of the LeishIF4G-4 region found to be essential for binding of LeishIF4E-3, as generated by the PsiPred server. Alpha helices (H) are shown as purple cylinders, and the confidence of the prediction is shown above (higher and darker blue columns represent higher confidence). Predictions are shown for the wild type sequence, as well as for the L26A and L26P mutants. (D) Mutational analysis of L26 in LeishIF4G-4 using a yeast two-hybrid assay. The assays were performed as described in B. The AD sequence was fused to the LeishIF4G-4-N (1–78) fragment carrying the L26A and L26P mutations. The BD sequence was fused to LeishIF4E-3. The yeast growth on plates is shown in Figure S2 and expression of the exogenous proteins in the transfected yeast strains is shown in Figure S1. (E) Transgenic L. amazonensis cells expressing SBP-tagged LeishIF4G-4 or LeishIF4G-4(L26P) were grown under normal conditions. Following washes and cell disruption, protein complexes were affinity-purified over streptavidin-sepharose, separated by SDS-PAGE and immunoblotted with antibodies against LeishIF4G-4 or LeishIF4E-3. The lanes were loaded with 1% [supernatant (S), flow through (F)] or with 20% [wash (W), elution (E)] of the total experimental sample.](/cms/asset/a4ab007d-4a8a-441a-98d2-0a322f902aba/krnb_a_10922709_f0009.gif)