Figures & data
Figure 1. (A) The NEAT1 long non-coding RNA (lncRNA) is an RNA polymerase II transcript with unusual features. Schematics of the NEAT1 genomic locus and NEAT1 transcripts are shown. Chromosome 11 is shown with the chromosome bands seen on Giemsa-stained chromosomes. The position of the chromosome locus can be estimated according to the numbers below. FRDM8 and SLC25A45 are the protein-coding genes located adjacent to NEAT1. The arrows indicate the directions of transcription. NEAT1_1 and NEAT1_2 possess distinct 3′-terminal structures. The triple-helix structure (red line) stabilizes NEAT1_2. The tRNA-like structure (gray line) is recognized by RNase P to create the 3′-end of NEAT1_2. (B) NEAT1 lncRNA (green) and PSPC1 (magenta) are localized to nuclear paraspeckles, which appear as bright nuclear foci in the right photograph. Nuclear DNA was stained with DAPI (blue). Scale bar is 10 μm.
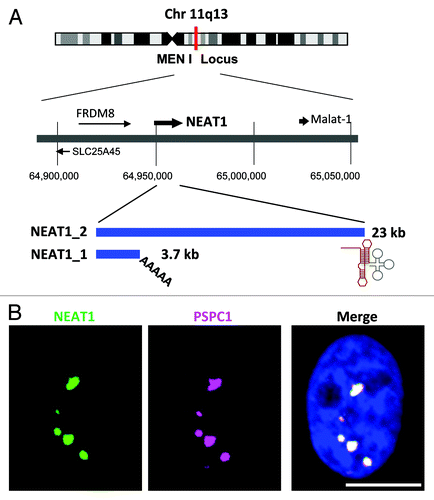
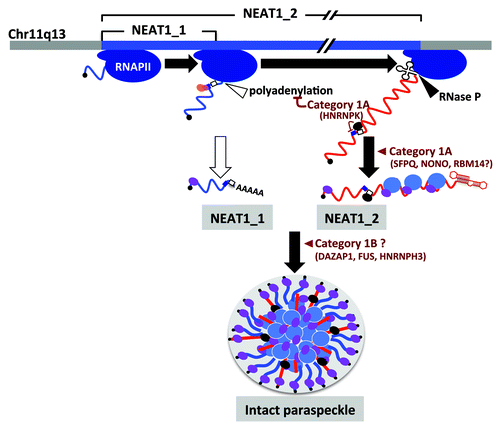
Figure 2. Paraspeckle formation proceeds during NEAT1 lncRNA biogenesis. NEAT1_1 and NEAT1_2 are shown by blue and red lines, respectively. The steps in paraspeckle formation in which essential PSPs (categories 1A and 1B) participate are shown. SFPQ (blue oval) and NONO (purple oval) preferentially bind to NEAT1_2. HNRNPK (black oval) binds to the pyrimidine stretch (open square) and arrests CFIm (pink and orange ovals)-dependent NEAT1_1 polyadenylation (CFIm-binding site cluster is shown by dark blue square).