Figures & data
Figure 1. Structure of the Spisula 45S rRNA cassette. Red segments represent transcribed spacer regions, blue segments correspond with mature rRNA subunits. Length of the 45S cassette is indicated in the scale bar, below. Gray bars at the top of the diagram indicate riboprobe target domains for the in situ hybridizations shown in and . Note that the 5′-ETS was targeted with two probes, one of approximately 1,000 nt, and one of 101 nt. The full nucleotide sequence of the Spisula 45S cassette is available under GenBank accession JN196041.
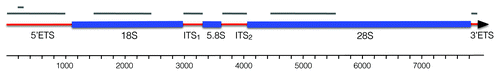
Figure 2. Localization of pre-rRNA subdomains in the unactivated oocyte. DIC images (top row) of unactivated oocytes and corresponding in situ hybridizations (bottom row). The 5′-ETS, ITS-1 and ITS-2 are strongly concentrated in the nucleolinus. No signal was detected using a riboprobe for the 3′-ETS. 18S and 28S rRNA show a granular pattern and are distributed throughout the cytoplasm as would be expected for ribosomes. Note that the 18S and 28S hybridization micrographs in this figure are taken slightly above the DIC focal plane so that all pertinent structures are visible. Corresponding opposite (sense) strand probes were used in each case as a negative control and yielded no signal above background. One example for each of the two observed labeling patterns (transcribed spacer and rRNA subunits) is shown in the last two pairs of images on the far right. Arrows indicate position of the nucleolinus. Three arrowheads in each of the DIC panels are used to delineate the boundaries of the nucleolus, which are difficult to see in some images.
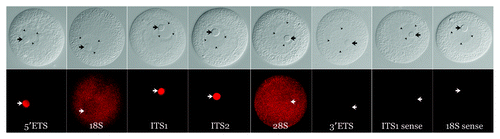
Figure 3. Presence of structural RNA in the procentrosome. Meiotic centrosomes in Spisula oocytes form in the cytoplasm 4‒5 min post-fertilization. They contain the universal centrosomal protein, γ-tubulin. Prior to 4 min, developing procentrosomes express the nucleolinar marker protein, nucleolinin, which is then co-expressed in the centrosome with γ-tubulin (B and C). Structural RNAs from the nucleolinus in the form of rRNA transcribed spacer sequences are present in these nucleolinin-containing procentrosomes (D and E). These particular RNAs are dissipated after γ-tubulin recruitment to the structure, while other transcripts (not necessarily associated with the nucleolinus) persist.Citation31-Citation33,Citation35 (A–C) shows an oocyte 8 min post-insemination co-labeled by indirect immunofluorescence for the centrosomal/procentrosomal marker nucleolinin and by direct immunofluorescence with a fluorophore-conjugated antibody for the centrosomal marker, γ-tubulin. (D–F) shows an oocyte at 2 min post-insemination co-labeled by indirect immunofluorescence with an antibody for the procentrosomal/centrosomal marker nucleolinin and a DIG-UTP riboprobe for the 5′-ETS imaged by direct fluorescence with a sheep-anti-DIG primary antibody.
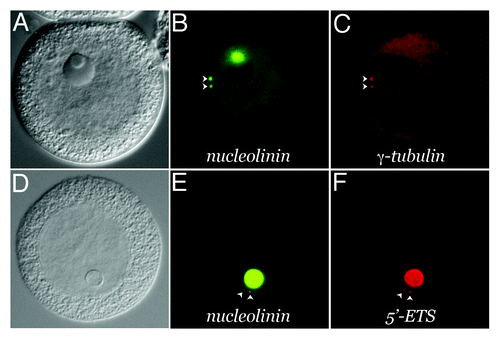
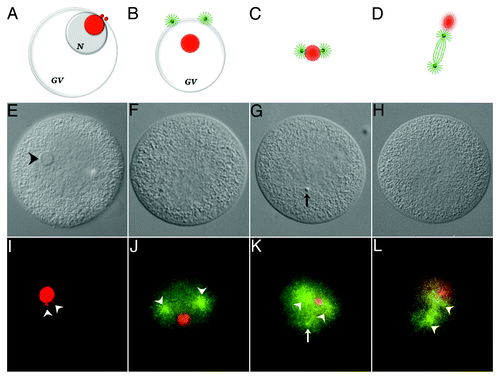
Figure 4. Association of 5′-ETS RNA with meiotic spindle components. The diagram shown in ) (A‒D) are not drawn to scale. Chromosomes and sperm components (nucleus and asters) are excluded for clarity. Nucleolinar RNA is colored red. Centrosomes, asters and spindle microtubules are colored green. GV, germinal vesicle; N, nucleolus. A, configuration up to approximately 8 min post-insemination. The nucleolinus is still intact. Procentrosomes and centrosomes hover at the periphery of the nucleolinar RNA patch before separating from each other and forming asters (B), which are first seen at the cytoplasmic edge of the collapsing GV (also see ). As the nucleolinus dissipates, a concentrated patch of RNA remains. By 12 min post-insemination, nucleolinar RNA and centrosomes (with growing spindle and astral arrays) remain closely associated (C). The nucleolinar RNA patch can be found between the developing poles, though not always arranged linearly. As the spindle begins to elongate, the nucleolinar RNA patch becomes smaller and more diffused around the edges, taking a position near one pole (D). (E‒H), DIC micrographs of oocytes fixed within the time blocks diagrammed above. (I–L), fluorescent images of the cells in (E–H) showing the distribution of 5′-ETS (red) and α-tubulin (green). Arrowheads in (I) highlight two 5′-ETS foci. Note that spindle integrity and α-tubulin antigenicity in these samples are severely compromised due to harsh hybridization conditions, including proteinase-K treatment and high temperature. Images of α-tubulin (microtubules) distribution in (J–L) are therefore of low quality, but nevertheless mark the vicinity of the asters and two half-spindles (arrowheads in J–L). A third astral array can be observed in some images (arrow in K). They are associated with the sperm nucleus, which is visible with bis-benzimide stain and often by DIC (arrow in G).