Figures & data
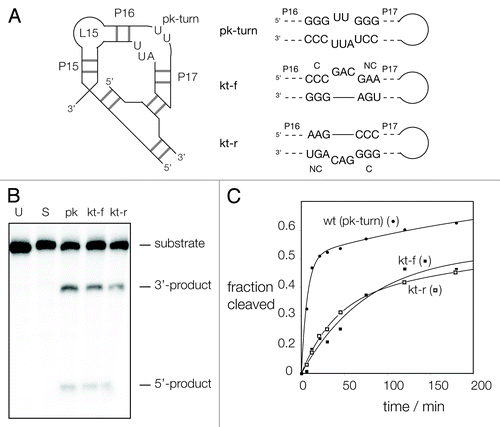
Figure 1. Sequences of pk and k-turn motifs in RNA. (A) The sequences of the T. tengcongensis SAM-I riboswitch k-turn and the T. maritima RNase P pk-turn. The standard nucleotide nomenclature is shown for the k-turn,Citation17 while the nucleotide numbering in the pk-turn follows that used in.Citation16 (B) A scheme showing the secondary structure of the pk-turn in RNase P. The arrows are deduced hydrogen bonds. Note that there are no hydrogen bonds between the strands. (C) Parallel-eye stereoscopic views of the individual structures of the two strands of the pk-turn taken from the crystal structure of T. maritima RNase P (PDB code 3Q1Q).Citation22 Deduced hydrogen bonds are indicated by gray, broken lines. (D) Superposition of the structures of the T. maritima RNase P pk-turn (loops cyan, basepairs green) (PDB code 3Q1Q) and the T. tengcongensis SAM-I riboswitch k-turn (loop magenta, basepairs gray) (PDB entry 3GX5).
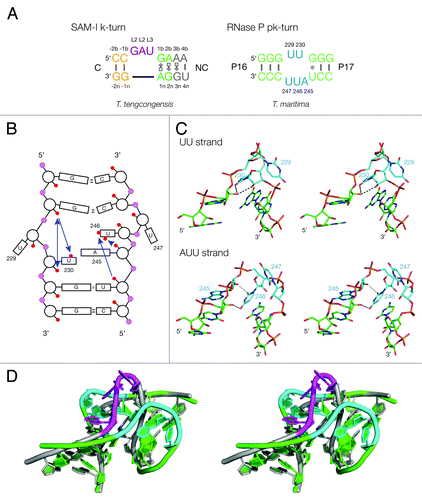
Figure 2. Analysis of ion-induced structural change in duplex RNA-containing a pk-turn studied by fluorescence resonance energy transfer in the steady-state. The pk-turn was centrally located (boxed in the sequence shown) within a 22 bp duplex of RNA terminally 5′-labeled with fluorescein (F, donor) and Cy3 (Cy, acceptor) fluorophores. FRET efficiency was measured in 90 mM Tris.borate (pH 8.3) with addition of MgCl2 to the indicated concentrations. The data are plotted as FRET efficiency (EFRET) as a function of Mg2+ concentration. Data for the SAM-I riboswitch k-turn are plotted (broken line) as the best fit to a two-state ion-induced transition, taken from.Citation13 Note that there is no increase in EFRET for the pk-turn-containing RNA at any Mg2+ concentration.
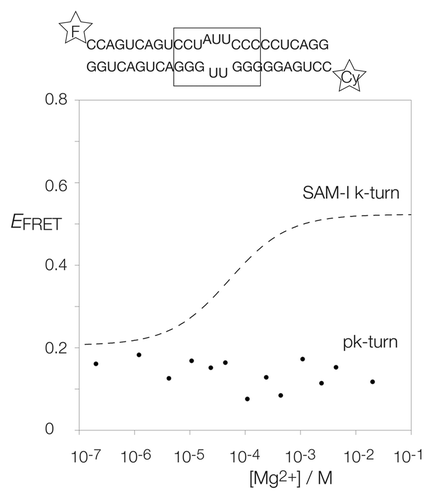
Figure 3. Analysis of the distribution of fluorescein fluorescent lifetime distributions for pk-turn and k-turn-containing RNA as a function of the presence or absence of Mg2+ ions. The same fluorescein, Cy3 pk-turn-containing RNA and an equivalent species containing the H. marismortui Kt-7 were used in these experiments.
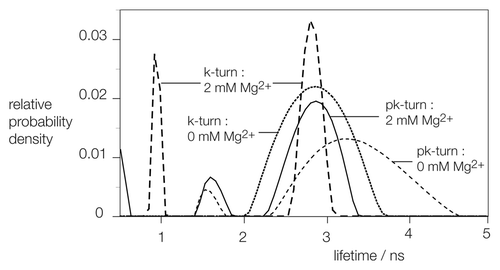
Figure 4. Replacement of the SAM-I riboswitch k-turn with the pk-turn. (A) A scheme shows the secondary structure and tertiary interaction of the SAM-I riboswitch, with the SAM-binding site indicated. The natural k-turn has been replaced by the pk turn in this structure. (B) Plots showing the results of microcalorimetric analysis of ligand binding to the SAM-I riboswitch containing the pk-turn. The upper panel shows the raw data for sequential injections of 2 µl volumes of a 5 µM solution of SAM into a 1.4 ml volume of 96 µM RNA solution in 50 mM HEPES (pH 7.5), 100 mM KCl, 10 mM MgCl2. This represents the differential of the total heat evolved for each SAM concentration (i.e., ∆H°). The lower panel presents the integrated heat data fitted to a single-site binding model (EquationEq. 1).
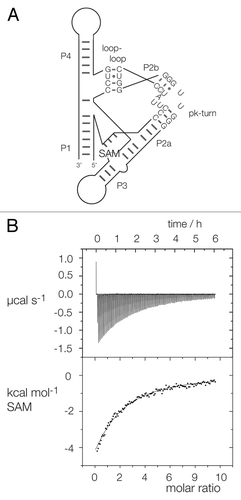
Table 1. Thermodynamic parameters for the SAM-I riboswitch
Figure 5. Ribozyme activity on RNase P in which the pk-turn has been replaced by a k-turn sequence in two orientations. (A) The secondary structure of the P15-P16-P17 section of RNase P showing the location of the pk-turn, and the tertiary interaction made by the terminal loop. The gray bars indicate basepairing, but the number of them is not significant. The ribozyme substrate binds at the loop L15. The sequence of the pk-turn and the forward and reverse oriented k-turns are shown. These are each located in RNase P in the same sense, i.e., P17 and its terminal loop is on the right-hand side as written. (B) Ribozyme activity of the natural and substituted forms of RNase P. Radioactive substrate (133 nt) and products (42 and 91 nt) were separated by gel electrophoresis following incubation with RNase P for 1 h. at 50°C. Substrate was loaded directly onto the gel without incubation (U), or after 1 h of incubation under assay conditions without added RNase P (S). The remaining samples were incubated for 1 h with RNase P, of native sequence including the pk-turn (pk), or with the pk-turn replaced by the SAM-I k-turn in the forward (kt-f) or reverse (kt-r) direction. Note that substrate is cleaved by all three ribozymes, at a slightly reduced level for the k-turn-containing forms. (C) Reaction progress plotted for the natural and substituted forms of RNase P. Symbols: natural ribozyme (containing the pk turn), closed circles; ribozyme containing the kt-f k-turn, closed squares; ribozyme containing the kt-r k-turn, open squares. The data are fitted to one or two exponential functions (lines).