Figures & data
Figure 1. The first A/T-hook of HMGA1 recognizes the apical stem structure of TAR. (A) Predicted secondary structures of 7SK L2 RNA and HIV-1 TAR. The putative HMGA1a-binding motif is labeled in green and the sites of interaction with Tat and Cyc T1 are highlighted in gray. (B) Predicted secondary structures of HIV-1 TAR mutants. The putative HMGA1a-binding site is labeled in green and mutated nucleotides are labeled in red. (C) Fam6-labeled HIV-1 TAR RNA was incubated with immunopurified HMGA1a-FLAG in presence of the 10-fold excess of the non-labeled TAR mutants shown in (B). The resulting complexes were analyzed in EMSAs compared with no competitor (-) and HIV-1 TAR wild-type. (D) Comparison of the predicted tertiary structures of the assumed HMGA1-binding motif of 7SK L2 and HIV-1 TAR. The tertiary structure was calculated using Assemble software.40 Adenine residues are labeled in blue, uracil residues in green, guanine residues in red and cytosine residues in yellow. (E) EMSAs with Fam6-labeled HIV-1 TAR and immunopurified HMGA1a-FLAG mutants. HIV-1 TAR was incubated with equal amounts (upper panel) of immunopurified mutants of each HMGA1a A/T-hook alone (HMGA1a ΔA/T1-3) and the resulting complexes were compared in EMSAs to HMGA1a-FLAG wild-type and the triple mutant HMGA1a 3xΔA/T-FLAG (lower panel).
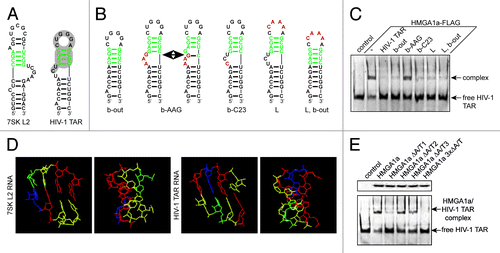
Figure 2. HMGA1a specifically interacts with HIV-1 TAR. (A) Electrophoretic mobility shift assays with Fam6-labeled HIV-1 TAR and immunopurified HMGA1a-FLAG. The TAR-binding activity of HMGA1a-FLAG (-) was compared with that of a mutant lacking DNA-binding activity (HMGA1a 3xΔA/T-FLAG) and TAR alone (control). Increasing amounts of non-labeled TAR and 7SK L2 RNA compete with labeled TAR for HMGA1a binding. Incubation with anti-FLAG antibody is capable of supershifting the HMGA1a-FLAG/TAR complex. (B) Fam6-labeled 7SK L2 RNA was incubated with immunopurified HMGA1a-FLAG in presence of increasing amounts of non-labeled HIV-1 TAR. The resulting complexes were analyzed in EMSAs. (C) Kd determination of HMGA1a complexes with HIV-1 TAR RNA and 7SK L2 RNA using microscale thermophoresis (MST). Fam6-labeled HIV-1 TAR or 7SK L2 RNA were incubated with increasing concentrations of immunopurified HMGA1a-FLAG compared with HMGA1a 3xΔA/T-FLAG and subsequently subjected to MST.
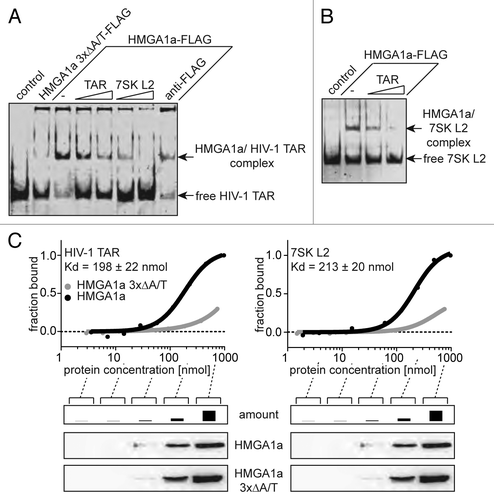
Figure 3. HMGA1 competes in vitro with HIV-1 Tat for TAR-binding. (A) HMGA1 and HIV-1 Tat compete for HIV-1 TAR binding. Fam6-labeled HIV-1 TAR was incubated with immunopurified HMGA1a-FLAG (HIV-1 Tat-FLAG) in presence of increasing amounts of immunopurified HIV-1 Tat-FLAG (HMGA1a-FLAG). The resulting complexes were separated by native PAGE in comparison to HIV-1 TAR RNA or HMGA1a-FLAG and Tat-FLAG alone. The Fam6-labeled RNA was visualized at UV light (upper part, left panel). Protein complexes were subjected to western blot analyses using anti-HIV-1 Tat antibody (middle part, left panel) and anti-FLAG antibody (lower part, left panel). The relative intensities of the signals were plotted using Image J software (right part). (B) Fam6-labeled 7SK L2 RNA was incubated with immunopurified HMGA1a-FLAG or Tat-FLAG and the resulting complexes were analyzed in EMSAs compared with 7SK L2 RNA alone (control). Protein complexes were subjected to western blot analyses using anti-HIV-1 Tat antibody.
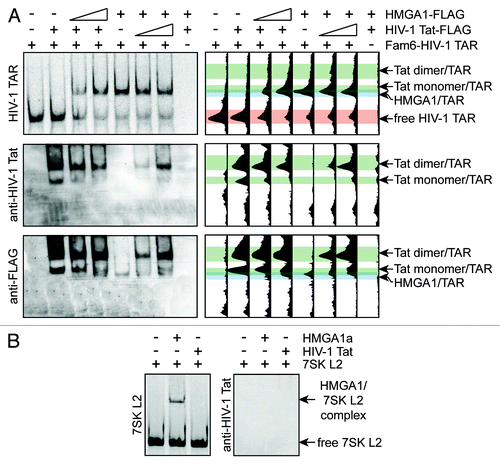
Figure 4. HMGA1 diminishes TAR-dependent HIV-1 transcription. (A) HeLa cells were subjected to a shRNA-mediated knockdown of HMGA1. The relative amount of HMGA1 mRNA was quantified using qantitative RT-PCR. The mRNA amount upon expression of a non-targeting control shRNA was abitrarily set to 1. (B) HMGA1-FLAG fusion protein was overexpressed in HeLa cells compared with mock transfected cells. HMGA1 amounts were quantified by western blot analyses using anti-FLAG and anti-HMGA1 antibodies. Protein amounts were quantified from anti-HMGA1 western blot signals using ImageJ software. (C) The 7SK L2 substructure (EBER2 7SK L2) was overexpressed in HeLa cells and compared with an overexpression of the EBER2 backbone (EBER2-Lb). The expression levels of both constructs were analyzed in quantitiative RT-PCR. β-actin was used as housekeeping gene and the levels of EBER2-Lb RNA were abitrarily set to 1. (D) Cells upon knockdown or overexpression of HMGA1 or overexpression of 7SK L2 (A–C) were either cotransfected with a mock vector or a HIV-1 Tat expression vector. After 24 h, they were transfected with either a wild-type HIV-1 LTR-driven luciferase reporter construct or a TAR-lacking HIV-1 LTR-driven luciferase reporter construct. Another 24 h later, relative promoter activity was assessed by measuring the luciferase activitiy. The promoter activity upon control shRNA, mock transfection and EBER2 backbone expression in the absence of Tat was abitrarily set to 1. (E) The HIV-1 promoter activity in the absence of Tat (D) was correlated with the relative amounts of HMGA1. (F) as in (E), but for the HIV-1 promoter activity in the presence of Tat.
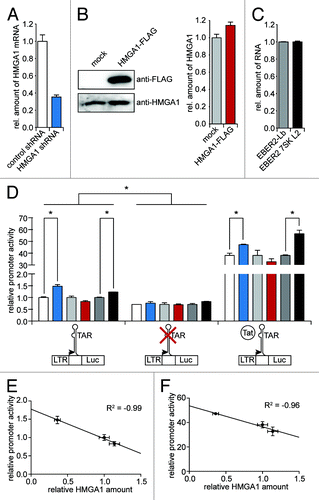
Figure 5. Model of the HMGA1-repressive role during HIV-1 transcription. (A) TAR-bound HMGA1 competes with HIV-1 Tat for TAR-binding. Tat targets P-TEFb from the promoter-bound inactive 7SK/P-TEFb snRNP, leading to the ejection of inactivating HEXIM1 and 7SK RNA, which subsequently competes with TAR for HMGA1 binding. That way, 7SK RNA facilitates the Tat/TAR interaction leading to the recruitment of activated P-TEFb to the paused RNA Polymerase II (RNA Pol II) to subsequently start efficient viral transcription. (B) In the absence of Tat, TAR-bound HMGA1 prevents the binding of transcription activating cellular co-factors to TAR (e.g., YB-1 or Pur-α). 7SK RNA competes with TAR vor HMGA1 binding and that way facilitates the TAR-binding of the cellular co-factors in order to activate basal viral transcription.