Figures & data
Figure 1. Sites of m6A are pervasive across the genome. This shows a circos plot of all m6A peaks for humans, with HEK293T cell peaks from Meyer et al. (in red, inner circle) and HepG2 from Dominissini et al. (in blue, inner-most circle). Peaks were found in over 10,000 genes and in all chromosomes (outer sections, varying colors).
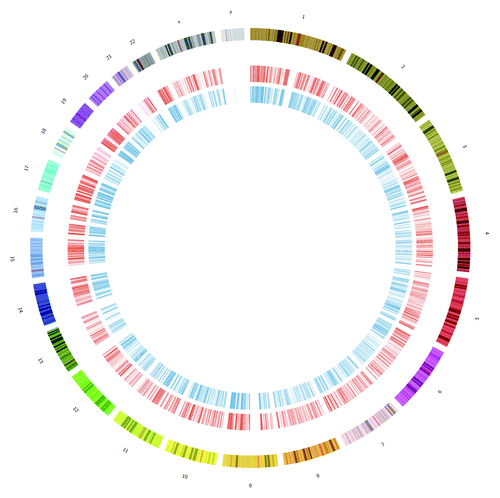
Figure 2. Effects of m6A on RNA function. We hypothesize that the sites of m6A (blue) will prevent the N-6 de-amination that occurs in RNA editing (red), where adenosine (A) gets converted into inosine (I) then guanosine (G). These sites may then have many roles in the function of RNA, from splicing to translation changes (blue).