Figures & data
Figure 1. Genomic location and context of lncRNAs. Protein-coding genes and their exons are represented by blue color, while lncRNAs and their exons are represented by red color. Panels are mainly based on lncRNA location annotation of GENCODE.Citation4 (A) Intergenic lncRNA, transcribed intergenically from both strands. (B) Intronic lncRNA, transcribed entirely from introns of protein-coding genes. (C) Sense lncRNA, transcribed from the sense strand of protein-coding genes and contain exons from protein-coding genes, overlapping with part of protein-coding genes or covering the entire sequence of a protein-coding gene through an intron. (D) Antisense lncRNA, transcribed from the antisense strand of protein-coding genes, overlapping with exonic or intronic regions or covering the entire protein-coding sequence through an intron.
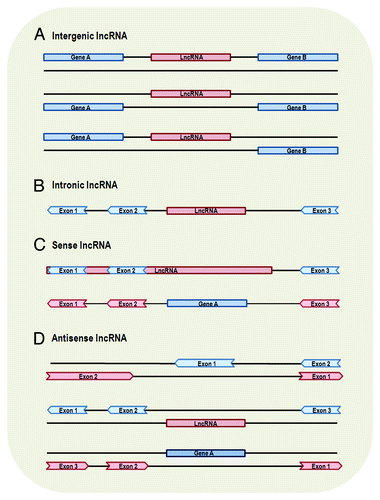
Table 1. Functional mechanisms and genomic locations of lncRNAs
Figure 2. Functional mechanisms of lncRNAs. LncRNA is represented by a letter “L” and a number appended. (A) Transcriptional regulation: We listed examples of cis-lncRNA (L1 and L3) and trans-lncRNA (L2). L1 is transcribed from the promoter region of gene A and its binding to promoter of gene A blocks the binding of transcription factors, thus affecting transcription initiation of gene A. L3 functions to modify chromatin protein in its vicinity through recruiting the complex of PRC2. L2 influences transcription of gene B from a distant region through interaction with transcription factor or RNA polymerase. Therefore, L1 and L2 also function through transcriptional interference, whereas L3 functions through chromatin modification. (B) Post-transcriptional regulation: L1, L2 and L3 all influence gene splicing. Specifically, L1 binds to intronic area to inhibit binding of splicing factor, L2 functions to modulate the pool of modified (such as phosphorylation) splicing factor and L3 binds to splicing factor to block spliceosomal complex formation. L4 interacts with translational factors to inhibit translation. L5 and L6 are two examples of ceRNAs, which interact directly or indirectly with miRNAs. L5 binds to miRNA and, thus, inhibits the binding of miRNA to the 3′ UTR of target mRNA. L6 binds to the 3′ UTR of target mRNA, which also blocks the binding of miRNA to the target gene. L7 serves as natural antisense inhibitor to promote degradation of mRNA. (C) Other functional mechanisms. L1 is involved in protein transportation and L2 binds to Dicer to influence RNA interference.
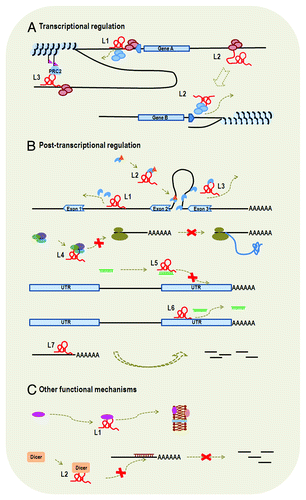
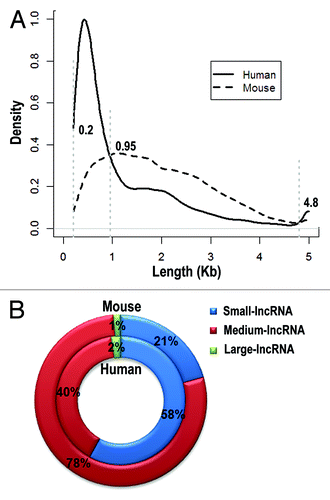
Figure 3. Length distribution of lncRNAs in human and mouse. LncRNAs are divided into three groups based on their length distribution: small-lncRNA (200~950 nt), medium-lncRNA (950~4,800 nt) and large-lncRNA (4,800 nt~). Density distributions of lncRNA length are shown in (A) and percentages of three lncRNA groups are depicted in (B).