Figures & data
Figure 1. Hfq/DsrA requirement at 24 °C and 37 °C. The steady-state levels of RpoS were determined by quantitative western blotting in the E. coli hfq- strain JW4130 harboring the control plasmid pACYC184 (lanes 1 and 3) and plasmid pAHfq, encoding Hfq (lanes 2 and 4), respectively. The strains were grown to early log phase (OD600 of 0.4) at 24 °C (lanes 1 and 2) or 37 °C (lanes 3 and 4). Equal amounts of cellular protein were loaded onto the SDS-polyacrylamide gel. Immunodetection of RpoS, ribosomal protein L14 (loading control), and Hfq as well as the detection of DsrA and 5S rRNA (loading control) by northern blot analysis was performed as described in the Materials and Methods. Only the relevant parts of the immunoblots and the autoradiographs are shown.
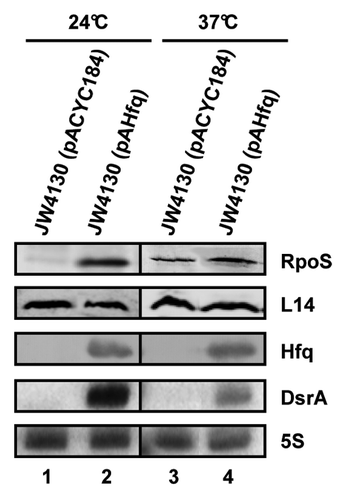
Figure 2. DsrA·rpoS duplex formation does not lead to efficient translation of rpoS mRNA in the absence of Hfq. (A) Immunodetection of RpoS, L9 ribosomal protein (loading control), and Hfq protein and detection of DsrA and 5S rRNA in the hfq- strain JW4130 harboring plasmid pNM12 (control; lane 1), pNM13 (encoding DsrA; lane 2), pACYC184 (control; lane 3), and pAHfq (encoding hfq; lane 4), respectively. The proteins and RNAs were visualized as described in the legend to . Only the relevant sections of the immunoblots and autoradiographs are shown. (B) Lanes 1–4, primer extension analysis of total RNA isolated from strains JW4130(pNM12), JW4130(pNM13), JW4130(pACYC184), and JW4130(pAHfq), respectively. The primer extension (PE) signals for rpoS mRNA isolated from the different strains are shown on top. The RNase III-mediated cleavage signals in rpoS mRNA are marked by arrows (G-112 and A-109). U, A, C, G, sequencing ladder.
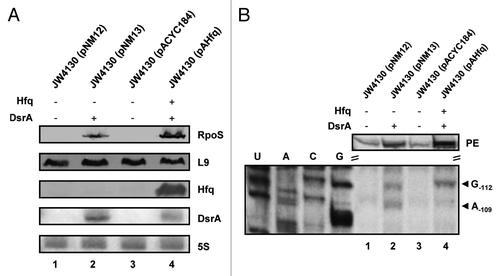
Figure 3. DsrA·rpoS duplex formation in the presence of HfqG29A does not result in efficient translation of rpoS mRNA at low temperature. (A) Immunodetection of RpoS, ribosomal protein L9, and Hfq in strains JW4130(pAHfq) (lane 1), JW4130(pAHfqG29A) (lane 2), and JW4130(pACYC184) (lane 3). DsrA and 5S rRNA were detected as described in the legend to . Only the relevant parts of the immunoblots and autoradiographs are shown. (B) Lanes 1–3, primer extension analysis of total RNA isolated from strains JW4130(pAHfq), JW4130(pAHfqG29A), and JW4130(pACYC184), respectively. The primer extension (PE) signals for rpoS mRNA isolated from the different strains are shown on top. The RNase III-mediated cleavage signals in rpoS mRNA are marked by arrows (G-112 and A-109). G, G sequencing ladder.
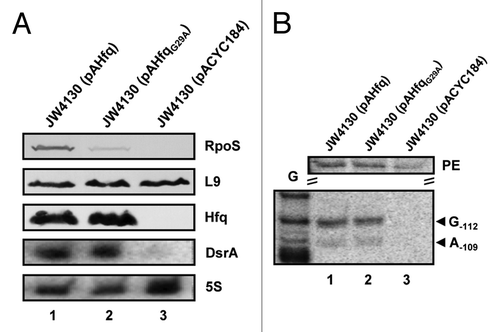
Figure 4. Structural probing of the translation initiation region of rpoS mRNA in complex with DsrA in the absence of Hfq and in the presence of Hfqwt or HfqG29A. First, rpoS mRNA was annealed to DsrA and to the [32P]-5′-end labeled primer. The annealing mix was cooled to 24 °C and divided into three parts followed by incubation for 10 min in the absence and presence of Hfqwt and HfqG29A, respectively. Then RNase T1 was added for 10 min followed by primer extension with AMV reverse transcriptase at 45 °C. The primer extension products were separated on a 8% polyacrylamide-8M urea gel and visualized by a PhosphorImager (Molecular Dynamics). The sequence of the immediate coding region is shown at the left. The start codon is marked by dots. RNase T1 cleavage 3′ of G residues and stop signals of the reverse transcriptase at A residues are indicated at the right. The numbering is given with regard to the A of the start codon (+1). Lane 1, primer extension without RNase T1 digestion. Lane 2–4, structural probing with RNase T1 in the absence of Hfq, in the presence of Hfqwt and in the presence of HfqG29A, respectively. U, A, C, G, sequencing ladder.
![Figure 4. Structural probing of the translation initiation region of rpoS mRNA in complex with DsrA in the absence of Hfq and in the presence of Hfqwt or HfqG29A. First, rpoS mRNA was annealed to DsrA and to the [32P]-5′-end labeled primer. The annealing mix was cooled to 24 °C and divided into three parts followed by incubation for 10 min in the absence and presence of Hfqwt and HfqG29A, respectively. Then RNase T1 was added for 10 min followed by primer extension with AMV reverse transcriptase at 45 °C. The primer extension products were separated on a 8% polyacrylamide-8M urea gel and visualized by a PhosphorImager (Molecular Dynamics). The sequence of the immediate coding region is shown at the left. The start codon is marked by dots. RNase T1 cleavage 3′ of G residues and stop signals of the reverse transcriptase at A residues are indicated at the right. The numbering is given with regard to the A of the start codon (+1). Lane 1, primer extension without RNase T1 digestion. Lane 2–4, structural probing with RNase T1 in the absence of Hfq, in the presence of Hfqwt and in the presence of HfqG29A, respectively. U, A, C, G, sequencing ladder.](/cms/asset/c42b5d08-f412-47ba-b4ea-626bcfaa87bc/krnb_a_10927100_f0004.gif)
Figure 5. Overexpression of csdA does not rescue rpoS translation in the presence of HfqG29A. Immunodetection of RpoS, CsdA, Hfq, and ribosomal protein L9 in strains JW4130(pAHfq;pProEx-Htb) (lane 1), JW4130(pAHfq;pCsdA) (lane 2), JW4130(pAHfqG29A; pProEx-Htb) (lane 3), and JW4130(pAHfqG29A;pCsdA) (lane 4), respectively. The proteins were visualized as described in the legend to . Only the relevant sections of the immunoblots are shown.
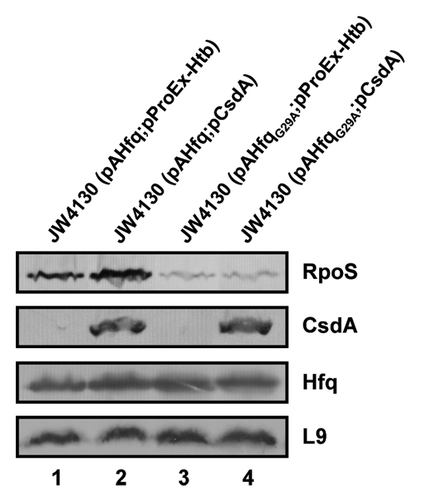
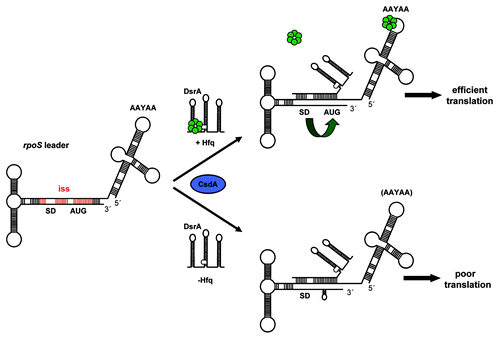
Figure 6. Working model for translational activation of rpoS mRNA at low temperature. The rbs of rpoS mRNA is masked by an iss (in red). In the presence of Hfq (upper scheme) the sRNA DsrA is bound and stabilized by Hfq.Citation4,Citation18,Citation24 CsdA (blue oval) is required for DsrA·rpoS duplex formation,Citation20 which leads to opening of the iss and in displacement of Hfq from DsrA.Citation26 Hfq bound to A-rich segments in the rpoS leader (AAYAA) promotes DsrA·rpoS annealingCitation24 and restructuring of rpoS mRNA into a translationally competent conformer (green bent arrow), which, in turn, permits efficient rpoS translation. In the absence of Hfq (lower scheme), DsrA·rpoS duplex formation occurs when dsrA is overexpressed. However, in the absence of Hfq, rpoS is poorly translated because the local secondary structure within the immediate coding region is not efficiently resolved. The Shine-Dalgarno (SD) sequence and start codon (AUG) in rpoS mRNA are highlighted, Hfq is shown in green.