Figures & data
Table 1. tRNA isodecoder genes in the hg19 reference human genome with scores ≥ 50 (from ref. Citation10)
Table 2. tRNA isodecoder genes in the hg19 reference human genome with scores < 50 (from ref. Citation10)
Figure 1. Coverage and sequence mismatch frequencies on selected tRNA genes. Deep sequencing data of DNA of individuals participating in the 1000-genomes project mapped on tRNA genes shows coverage along the gene and highlights sequence mismatches for each gene’s positions. All baseline sequences are from the corresponding tRNA gene sequences in the hg19 reference genome.Citation10 The coverage is determined by piling up all deep sequencing reads that map to the gene, and shown as vertical black bars. Sequence mismatch frequencies per position are marked by red triangles set at heights proportional to the frequencies: higher red bars indicate higher mismatch frequencies. Five highest mismatched columns are highlighted in green. Selected tRNAs are: (A) tRNALeu(CAA). The well-shaped coverage is due to the fact that only a few tRNALeu genes have introns at positions corresponding to the bottom of the well. (B) tRNATyr(GUA). Greater coverage at the 5′ region of the tRNA gene gives an indication of the degeneracy of that region among the isodecoders. (C) tRNAArg(ACG). (D) tRNACys(GCA).
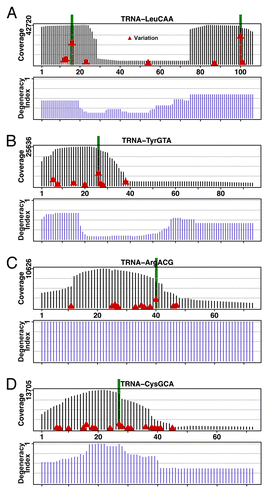
Figure 2. Distribution of new tRNA genes. All reference genome tRNA genes are separated into two groups according to their tRNAscan-SE scores to facilitate analysis. (A–C) Scores ≥50 listed in ; (D–E) Scores <50 listed in . (A) High-scoring tRNA group. Distribution of new isodecoder instances per isoacceptor family. The most frequently occurring new isodecoder codes for leucine (53%), followed by tyrosine (22%). Only those tRNAs with more than five new instances are shown. Unique new isodecoder instances found in >1% of the population is also shown. Each instance is tagged with a Roman number. The nucleotide positions are adjusted according to the standard tRNA nomenclature. The new genes indicated by a star were tested experimentally for their folding and in vivo charging and stability. (Inset) Distribution of new isodecoder instances among individuals. Over half (54%) of all individuals carry at least one new isodecoder. Most individuals (81%) carry only one or none new isodecoder, while few individuals have as many as 11 new tRNA isodecoders. (B) Sequence change for the 11 new genes that are present in > 1% population mapped onto the canonical tRNA structure. Roman numerals correspond to those of the previous panel. (C) Box-and-whisker plot showing the distribution of new isodecoder instances per ancestry codes. Thick horizontal bars indicate the median, while the bottom and top of the boxes the first and third quartiles of the distributions, respectively. (D) Low-scoring tRNA group. Distribution of new isodecoder instances per isoacceptor family. The most frequently occurring new isodecoder codes for cysteine (57%), followed by a suppressor tRNA (17%). Only those tRNAs with more than five new instances are shown. Unique new isodecoder instances found in > 1% of the population is also shown. Each instance is tagged with a Roman number. The nucleotide positions are adjusted according to the standard tRNA nomenclature. (E) Sequence change for the 13 new genes that are present in >1% population mapped onto the canonical tRNA structure.
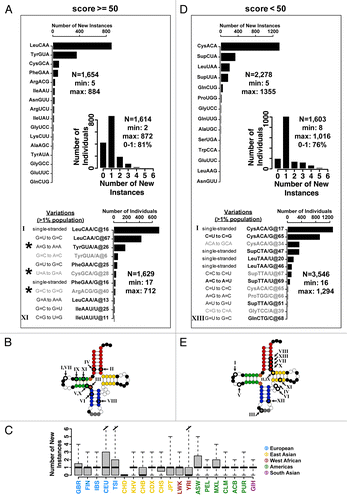
Figure 3. Population analysis of the most frequent new tRNA sequences among the high scoring tRNA group. (A) Sequence change mapped onto the canonical tRNA secondary structure for tRNALeu(CAA), the tRNA gene with the most abundant new instances. Square bubbles indicate the nucleotide substitution frequencies. Conserved nucleotides are shown in black, while the anticodon nucleotides in gray. Dotted lines show base paired positions. (B) Probability table conditioned on ancestry for position 16 of tRNALeu(CAA). The SoAs label stands for South Asian, Amer for Americas, WeAf for West Africa and Euro for European. (C) Sequence change mapped onto the canonical tRNA secondary structure for tRNATyr(GUA), the tRNA gene with the second most abundant new instances. (D) Probability table conditioned on ancestry for position 26 of tRNATyr(GUA). The SoAs label stands for South Asian, EaAs for East Asian. (E and F) Sequence changes in the anticodon stem in two other abundant new isodecoders. The sequence changes introduce a purine mismatch in the anticodon stem. (E) Position 40 of tRNAArg(ACG) occurring in 15.5% population. (F) Position 27 of tRNACys(GCA) occurring in 11.3% population.
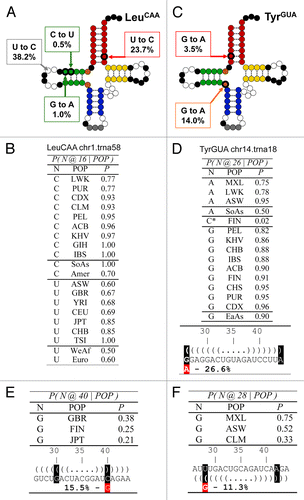
Figure 4. Structural mapping of pairs of tRNA isodecoder transcripts in vitro. Structural mapping of selected tRNA isodecoder pairs was performed by limited nuclease V1 and S1 digestion. Sequences in the context of standard tRNA secondary structure and mapping of tRNAArg(ACG) (A), tRNACys(GCA) (B), and tRNATyr(GUA) (C). Positions of RNA cuts were identified through alkaline hydrolysis (BH lane) and T1 ladder (T1 lane). The sequence change in the new tRNA isodecoder is indicated by red arrows and corresponds to the position of the red dot in gel. tRNA structural regions are indicated on the right side of the gel. (D) tRNA folding analysis by native gel electrophoresis. tRNA transcripts from T7 RNA polymerase reaction was directly loaded on 8% native gel containing 25 mM trisOAc, pH 7.5, 5 mM Mg(OAc)2. The amount of transcription reaction loaded in the left and right panels were 1 and 10 µl, respectively. Consistent with the structural mapping results, tRNATyr(GUA) variants fold in the same manner, whereas tRNAArg(ACG) variants fold very differently.
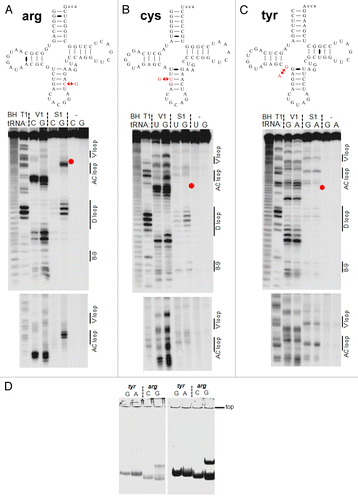
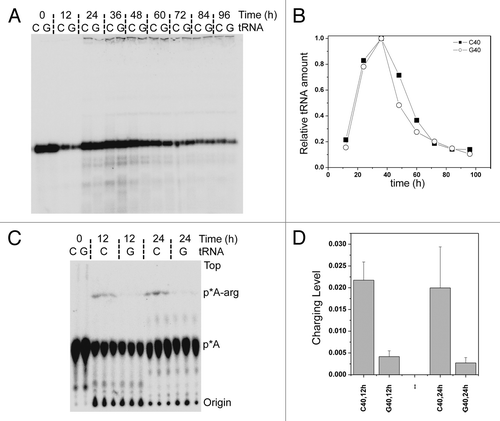
Figure 5. Stability and charging level of tRNAArg(ACG) isodecoder pair in vivo. (A) tRNA transcripts 32P-labeled at the terminal A76 were transfected into HeLa cells. Total RNA was isolated at designated time points, and equal amount of total RNA was loaded in each lane. tRNAArg(ACG) from the reference genome has C40 and is indicated as C lanes; the new isodecoder has G40 and is indicated as G lanes. (B) Quantitative analysis of the 32P-labeled tRNA bands in panel A. Data are normalized to the 36 h time point where it has the highest level of radioactivity. (C) tRNA transcripts 32P-labeled at the terminal A76 were transfected into HeLa cells. Total RNA under acidic conditions was isolated at designated time points. Equal amount of total RNA was digested with nuclease P1 and run on TLC. Different charging level of tRNA isodecoders at different time points analyzed by TLC. Free and acylated tRNA are indicated on the right side. (D) Quantitative analysis of the p*A and p*A-arg spots in panel A. The y-axis corresponds to the % of p*A-arg spot over p*A+p*A-arg signals.