Figures & data
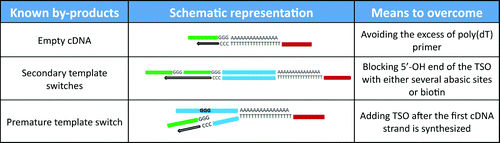
Figure 1. Schematic representation of cDNA preparation methods using a combination of poly(A) or poly(dA) tailing and template switching capacity of MMLV-RT.
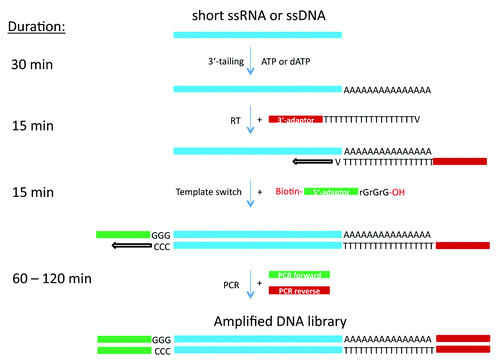
Figure 2. (A) The structure of cDNA prepared using adaptor sequences for the Illumina sequencing platform. Note, the absence of sequence complementarity between PCR primers and terminal adaptors allowing pre-amplification of the cDNA library without prior purification of the first cDNA strand. (B) Electropherogram obtained after 3% agarose gel electrophoresis of amplified DNA libraries obtained from 1 ng (or 5 pg) of either synthetic cel-miR-39 RNA or DNA molecules. The number of PCR amplification cycles and the concentration of reverse poly(dT) primer are indicated below each electropherogram. In addition, Agilent Bioanalyser (High Sensitivity DNA chips) and automated Sanger sequencing were used to estimate the purity of DNA libraries after the column purification step (upper chromatogram) and additional gel extraction step (lower chromatogram). Agilent Bioanalyser data and Sanger chromatograms shown only for 5 pg RNA and DNA inputs.
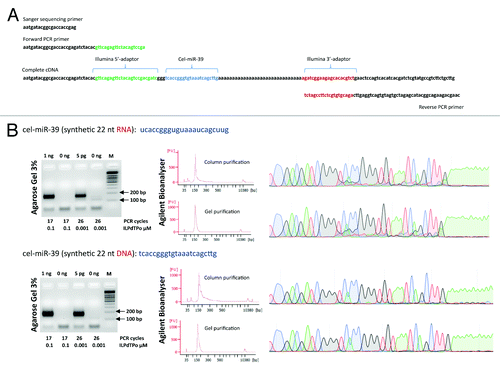
Figure 3. Nucleotides distribution of reads obtained after MiSeq Illumina sequencing of the libraries generated from: (A) Poly(A) enriched RNA (R10 and R5) and bisulfite converted DNA (B) from U2OS cells; (B) Human blood plasma circulating DNA (DI and DII) and circulating RNA (RI). Note, the significant bias toward reads containing 5′-terminal G nucleotides in the RNA samples. In addition, RNA molecules starting with A nucleotide are incorporated into the DNA library with lower efficiency. Note, no bias toward a 5′-terminal nucleotide was observed when DNA was taken as input. Note, the significantly lower content of dC nucleotide in the libraries generated from bisulfite-converted DNA.
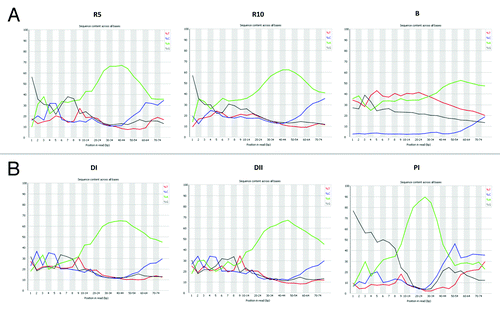
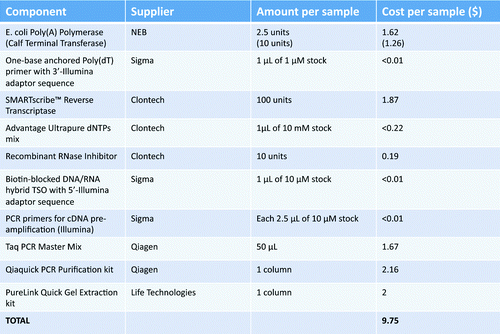
Table 2. Cost estimation of the consumables per sample of cDNA preparation according to the here presented protocol. Calculations include prices in US dollars without special discounts (as by end of 2013). Common consumables (e.g., laboratory plastic, gel electrophoresis reagents etc.) have not been included in the calculations.