Figures & data
Figure 1. Schematic representation of GFPendo, hpF and GFPcDNA constructs. P35S: CaMV 35S promoter; TNOS: nopaline synthase (NOS) terminator; GFP: green fluorescent protein; PRbc: tobacco RuBisCO promoter including 5′ UTR; TRbc: tobacco RuBisCO terminator including 3′ UTR; Intron 1 and 2: tobacco RuBisCO introns 1 and 2. The hybridization probes used for northern blot analysis () and the 182 bp and 134 bp regions analyzed by bisulfite sequencing (, S2 and S3) are indicated by black bars.
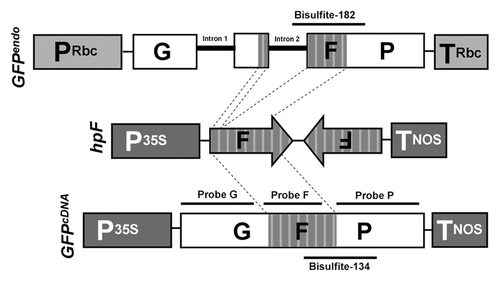
Figure 2. GFP monitoring of hpF-agroinfiltrated GFPendo and GFPcDNAN. benthamiana plants. The development of RNA silencing was monitored at local levels (4 and 8 dpi) and systemic levels (21 and 46 dpi).
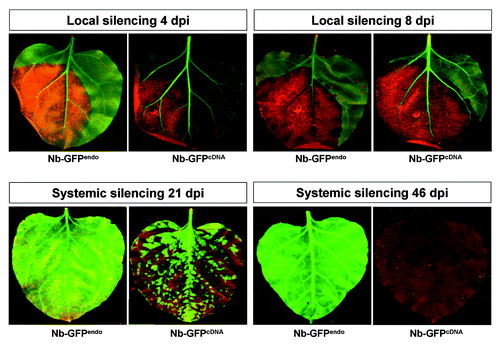
Figure 3. Northern blot analysis of RNA samples of hpF-agroinfiltrated GFPendo and GFPcDNAN. benthamiana plants. Large and small RNA analysis was performed at local levels (4 and 8 dpi) and systemic levels (21 dpi). The regions corresponding to hybridization probes are indicated in . Ethidium bromide staining and miR159 accumulation served as loading controls. 'Mock' corresponds to 10 mM MgCl2 infiltration.
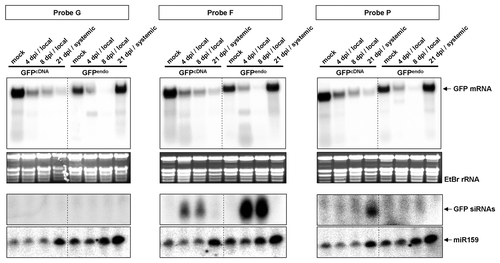
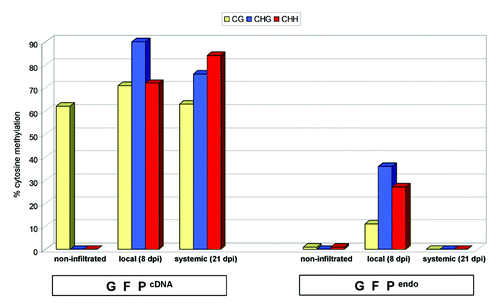
Figure 4. Histogram representing bisulfite sequencing data from GFPcDNA and GFPendo. Non-agroinfiltrated, 8 dpi local and 21 dpi systemic tissue was used for analysis. 10–15 clones were sequenced and data were analyzed with CyMate software.Citation68 Histogram represents the data presented in Fig. S2 and S3. In non-agroinfiltrated GFPcDNA, transgenerationally maintained CG methylation was detected, in agreement with previous observations.Citation41,Citation70