Figures & data
Figure 1. General scheme of mRNA localization. The formation of a splicing-dependent stem-loop and involvement of the EJC is seen in the localization of the transcript oskar in Drosophila embryos. After splicing, mRNAs are exported from the nucleus. Those containing mRNA localization signals, in this case, the stem-loop, are recognized by trans-factors that interact with the localization signal. The trans-factor is integrated into a transport complex that uses motor proteins to move the RNA to its destination. Localized RNAs are often kept in a translationally-repressed state until they reach their destination, whereupon they are released and may be translated to give a localized pool of protein product.
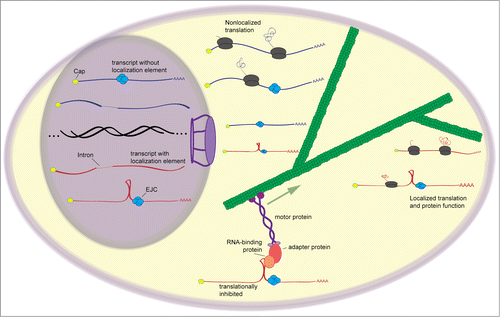
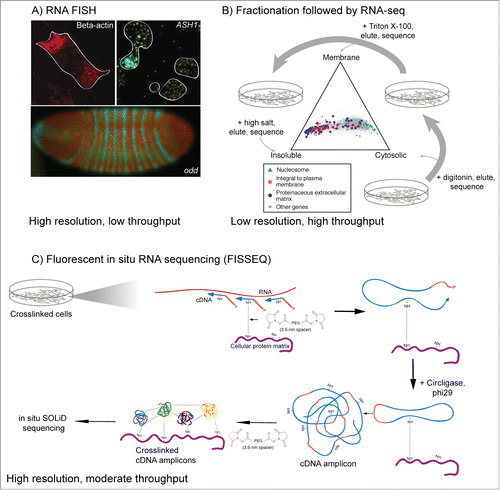
Figure 2. Classical and genomic approaches for studying RNA localization. (A) RNA FISH images from three systems. RNA encoding β-actin is localized to the leading edges of chicken fibroblasts,Citation124 while ASH1 mRNA localizes to the bud tip of S. cerevisiae during mating type switching.Citation125 RNA encoding odd and the resulting protein product accumulate in distinct segments in the developing Drosophila embryo and are important for proper development and morphology.Citation35 (B) Fractionation of cells followed by high-throughput sequencing can give a more generalized view of RNA localization.Citation21 Here, mouse C2C12 myoblasts were treated first with digitonin, which gently lyses cells but keeps membranes generally intact. Cytoplasmic RNA is collected and the cells are then treated with Triton X-100 to solubilize membranes. Membrane-associated RNA is collected and the resulting sample is treated with high salt to elute RNA associated with insoluble cell components. RNA from each fraction is then analyzed by RNA-seq. The location of activity of the protein encoded by each RNA generally match with the location of the RNA. (C) Method overview of FISSEQ.Citation46 Cellular structures are crosslinked followed by in situ reverse transcription of the crosslinked RNA using random primers and modified nucleotides that allow further crosslinking. The resulting cDNA is crosslinked to cellular structures and the circularized and amplified using rolling circle amplification. cDNA amplicons are then crosslinked to cellular structures and each other and sequenced in situ using SOLiD sequencing.