Figures & data
Figure 1 Altered Expression of SRrp86. Knockdown of SRrp86 was achieved by transfecting siRNAs using TransIT TKO (Mirus) for four consecutive days. Overexpression was achieved by transfecting a rat SRrp86 expression vector using TransIT LT1 (Mirus). Protein samples were harvested and separated on 8% SDS-PAGE gels, transferred and probed with either a rabbit polyclonal antibody to SRrp86 or an antibody to α-tubulin. (A) SRrp86 knockdown in HeLa cells. (B) SRrp86 knockdown in H1299 cells. (C) SRrp86 overexpression in HeLa cells. (D) SRrp86 overexpression in H1299 cells.
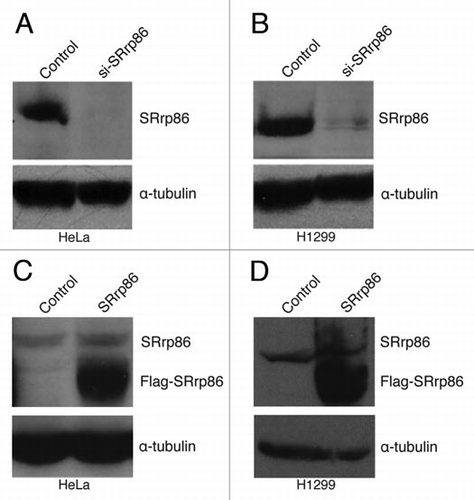
Figure 2 Probe design for the SpliceArray. Six probes were used to detect specific splicing events. Two of the probes, T and F, are designed to detect invariant mRNA regions to control for changes in expression levels. If mRNA detection with these probes changes across conditions, the changes are likely to reflect transcriptional effects and not altered splicing. Probes B, C, D and E span exon-exon junctions or reside entirely within variable exon sequences. Changes in mRNA levels detected with these probes relative to the invariant T and F probes indicate a change in splicing pattern.
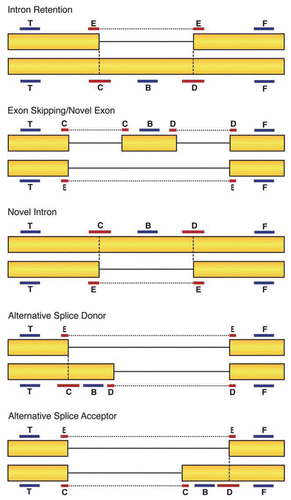
Figure 3 SRrp86 regulates an alternative splicing event in IκBβ and affects NFκB signaling. (A) The SpliceArray detected an alternative intron inclusion event between exons 5 and 6 of the IκBβ gene (NFKBIB). Under normal conditions both isoforms are expressed, but overexpression of SRrp86 promotes splicing of the intron and production of isoform 1. (B) Western blots were performed to detect changes in isoform expression in H1299 cells. Knock down of SRrp86 resulted in increased isoform 2 expression relative to isoform 1. Overexpression of SRrp86 resulted in increased expression of isoform 1 relative to isoform 2. (C) Flow chart outlining the effect SRrp86 expression on luciferase expression. SRrp86 enhances isoform 1 expression which allows for increased levels of basal and TNFá-stimulated NFκB activity. Loss of SRrp86 expression results in decreased levels of isoform 1 while maintaining the repressive activity of isoform 2 with the overall result being decreased NFκB activity. (D) Luciferase assays were performed to determine the effect of changing SRrp86 levels on NFκB signaling. Changes in the ratio of the two IκBβ isoforms due to SRrp86 were monitored using a construct containing the firefly luciferase gene under the control of NFκB. Basal (−) and stimulated (+) levels of NFκB activity were determined after treatment of cells with TNFα for 6 hr. Error bars represent SEM; *represents p < 0.05 compared to unstimulated control and **represents p < 0.05 compared to stimulated control by paired, 2-tailed t-test; n = 4.
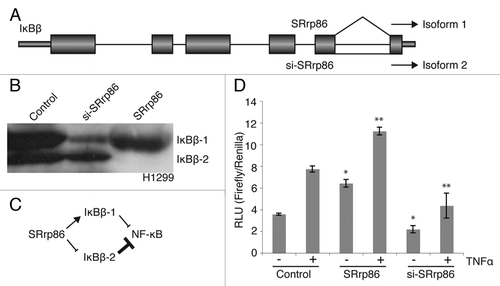
Figure 4 SRrp86 regulates a novel intron event in c-Jun. (A) The SpliceArray detected an aberrant splicing event in c-Jun upon overexpression of SRrp86. Normally, c-Jun produces a single exon mRNA but overexpression of SRrp86 caused removal of part of the 5′ UTR and 75% of the open reading frame. mRNAs resulting from this splicing event are incapable of expressing c-Jun protein. (B) Western blots were performed to analyze the effect of SRrp86 overexpression on c-Jun levels in H1299 cells. Overexpression of SRrp86 resulted in loss of c-Jun expression. Knockdown of SRrp86 had no effect. α-Tubulin was used as a loading control. (C) Flow chart outlining the effect of SRrp86 on luciferase expression using a reporter construct. Briefly, SRrp86 blocks c-Jun function and thus its binding to the AP-1-regulated promoter that drives luciferase expression. (D) Luciferase reporter assays were used to analyze the effect of the loss of c-Jun protein due to SRrp86 overexpression on AP-1 transcription factor complex activity. The AP-1 luciferase construct contains two copies of the AP-1 consensus DNA binding sequence in the pGL-3 promoter vector (Promega). Error bars represent SEM; *represents p < 0.01 by paired, 2-tailed t-test; n = 3.
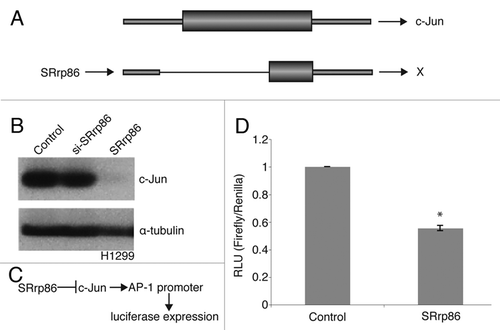
Table 1 SRrp86-regulated alternative splicing events in h1299 cells
Table 2 SRrp86-regulated alternative splicing events in heLa cells