Figures & data
Figure 1 Relative expression of Hsa21-derived miRNAs in human tissues. Mature human let-7c, miR-99a, miR-125b, miR-155 and miR-802 were quantified utilizing TaqMan microRNA assay kits specific for each Hsa21-derived miRNA (Applied Biosystems, Foster City, CA) as previously described.Citation40,Citation42 The expression values were normalized to RNU48 for each tissue. Relative gene expression was calculated as 2-(CTmiR-155-CT-RNU48). The relative abundance of each Hsa-21 derived miRNA is shown as a percentage of total combined expression of Hsa-21 derived miRNAs for each tissue investigated.
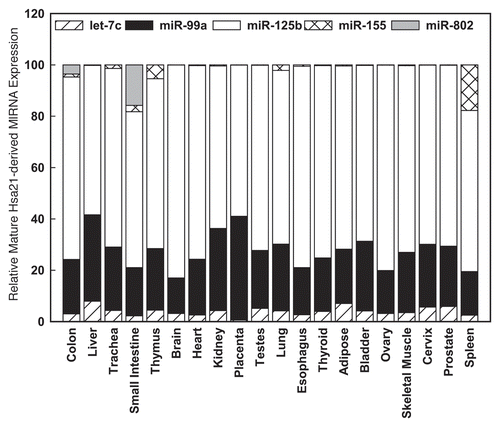
Figure 2 Hsa21-derived miRNAs are overexpressed in human DS pre-frontal cortex brain specimens. Mature Hsa21-derived miRNAs were quantified utilizing RT-PCR as previously described by our laboratoryCitation40,Citation42,Citation85,Citation86 using total RNA isolated from human fetal (18–22 weeks of gestation), child (1–8 years), adolescent (9–19 years old) and adult (20–50 years old) prefrontal cortex specimens from control and DS (age- and sex-matched, n = 3–5) samples. Gene expression was calculated relative to 18S rRNA and the data are expressed as percent over control (variation between control samples was never greater than 5%), which was assigned a value of 100%. The error bars represent the average ± S.E. of triplicate samples repeated in at least three independent experiments (*p < 0.01 DS vs. control).
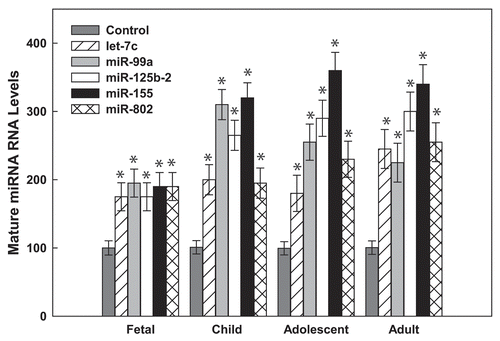
Figure 3 The human AT1R +1166 A/C SNP occurs in the miR-155-binding site. (A) Complementarity between miR-155 and the hAT1R 3′-UTR site targeted (70–90 bp downstream from the human AT1R stop codon). The +1166 A/C SNP corresponds to the nucleotide 86 bp downstream from the human AT1R stop codon (shown in bold print). The binding of miR-155 to the hAT1R 3′-UTR target site fulfills the requirement of a 7 bp seed sequence of complementarity at the miRNA 5′ end when the +1166 A-allele is expressed. (B) Complementarity between miR-155 and the human AT1R 3′-UTR harboring the +1166 C-allele. If the +1166 C-allele is expressed, the seed sequence requirement would not be met and, as a consequence, it would be expected that human AT1R expression would be elevated.
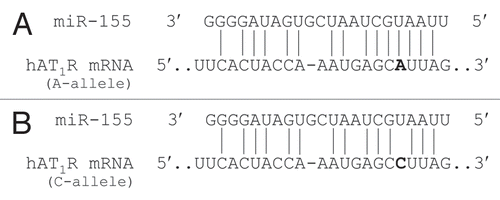
Table 1 Targetscan algorithm-predicted Hsa21-derived putative miRNA/mRNA targets