Figures & data
Figure 1 Steady-state levels of RpoS determined by quantitative western-blotting in the wt strain and the isogenic csdA mutant. The E. coli strains WJW45 (wt), the isogenic csdA deficient strain WJW45ΔcsdA (csdA−) as well as WJW45ΔcsdA harbouring plasmid pUCdeaDCitation38 (csdA−/pUCdeaD) were grown to early log phase (OD600 of 0.4) either at 37°C (lane 2–4) or at 24°C (lane 5–7), respectively. Cell extracts of the rpoS mutant strain RH90,Citation53 served as a negative control (lane 1). Equal amounts of total cellular proteins were loaded in each lane of the SDS-polyacrylamide gel. The RpoS protein was detected by immunological means as described in Materials and Methods. Only the relevant section of the immunoblot is shown.
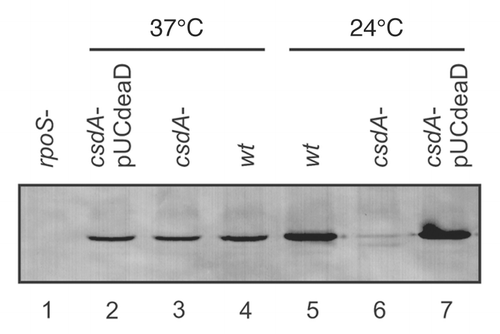
Figure 2 CsdA stimulates rpoS synthesis in vitro. 10 pmol of in vitro synthesized full length rpoS mRNA were translated using the PURESYSTEM. Translation of rpoS mRNA was performed in the absence (lanes 2–3) or in the presence of equimolar amounts of DsrA (lanes 4–7). Lane 1, no mRNA was added to the reaction mixture. Lane 2, equimolar amounts of the sRNA RyhB, which does not stimulate rpoS translation, was added. Lane 5 and 6, translation in the presence of 5 (+) and 10 pmol (++) CsdA, respectively. Lane 7, 10 pmol of heat-inactivated CsdA protein (ΔT°) was added to the reaction mixture. The protein samples were resolved on a 12% SDS-polyacrylamide gel. The signals were visualized by a PhosporImager (Molecular Dynamics). The experiment was performed in triplicate. Only the relevant section of one representative autoradiogram is shown. The position of the RpoS is indicated.
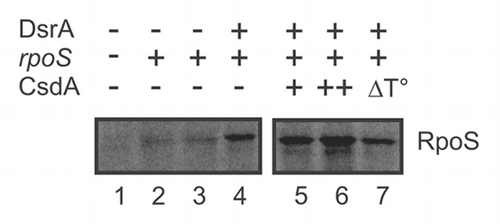
Figure 3 Primer extension analysis of total RNA isolated from the E. coli strain WJW45 (wt) and the csdA deficient strain WJW45ΔcsdA (csdA-) grown to an OD600 of 0.4 at 24°C. Top: The product of the RNase III cleavage within the rpoS leader (position G−112) accumulated only in the wild-type strain (lane 5) and was hardly detectable in the csdA deficient strain (lane 6). Lanes 1–4, sequencing ladder. The experiment was performed in triplicate. One representative autoradiograph is shown. Bottom: The DsrA RNA levels and 5S rRNA levels (loading control) of the wt and csdA-strain grown at 24°C were determined by northern-blot analysis as described in Materials and Methods. The Hfq protein levels in both strains were determined by western-blotting using equal amounts of total protein of the two strains at the time the cells were harvested for preparation of total RNA.
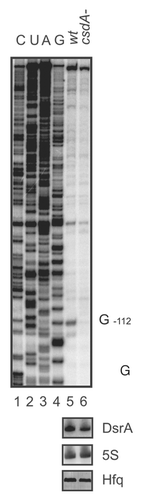
Figure 4 The interaction between CsdA and Hfq is RNA mediated. (A) Far western-blotting does not reveal an interaction between CsdA and Hfq. 25 pmol of CsdA-His6 (lane 1) or 25 pmol of CsdA-His6 (lanes 2) treated with micrococcal nuclease (MN) and 25 pmol of Hfq (lane 3, control) were separated by SDS-polyacrylamide gel electrophoresis and then electroblotted onto nitrocellulose membranes. After re-naturation of the proteins the membranes were incubated with Hfq (150 pmol Hfq as hexamer) as described in Materials and Methods. The CsdAHis6-Hfq complex was visualized by immuno-detection using anti-Hfq antibodies (lane 1), whereas no CsdA-Hfq complex could be detected with anti-Hfq antibodies after treatment with MN (lane 2). Lane 3, Hfq protein (control) was detected with anti-Hfq antibodies. (B) Co-immunoprecipitation assay with protein CsdA-His6 and Hfq using anti-Hfq antibodies. Lane 1, CsdA-His6 (10 pmol) was loaded as a positive control for the anti-His antibodies. 25 pmol of purified hexameric Hfq were incubated together with 50 pmol CsdA-His6 (lane 2) or 50 pmol CsdA-His6 treated with MN (lane 3) followed by immunoprecipitation using anti-Hfq antibodies bound to the Dynabeads (see Materials and Methods). The co-immunoprecipitated CsdA-His6 was detected on the western-blot with anti-His antibodies. Only the relevant sections of the immunoblots are shown.
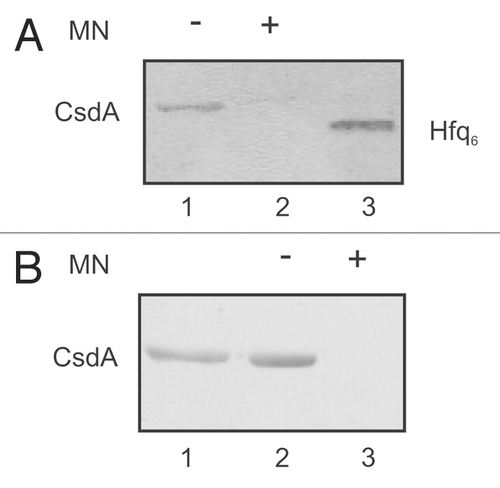
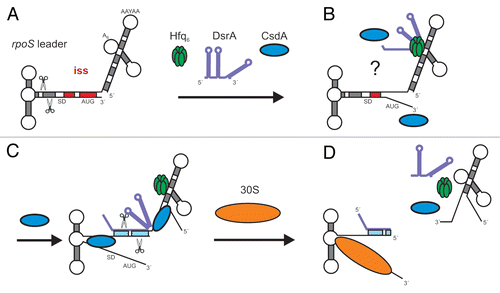
Figure 5 Model for translational activation of rpoS mRNA by DsrA, Hfq and CsdA. (A) At low temperature and during exponential growth RNase III cleavage occurs in the rpoS leader, which primes rapid decay of the mRNA.Citation25 As DsrA·rpoS annealing () and consequently rpoS translation () did not occur in the csdA- strain, intramolecular cleavage in the rpoS leader most likely accounts for the reduced steady state levels of rpoS mRNA in the latter when compared to the wild-type strain (see ). (B) Hfq-DsrA binding to A-rich segments (A6 and/or AAN(4)Citation30) in the rpoS leader occurs at low temperature when DsrA is expressedCitation30 (for clarity only one possible Hfq binding site is occupied by the Hfq-DsrA complex). CsdA aids in melting the iss and perhaps in unfolding of DsrA. (C) Annealing between DsrA and the rpoS mRNA occurs opposite of the rbs. (D) Translation of rpoS ensues and intermolecular RNase III cleavage of the DsrA·rpoS duplex (at G−112 in rpoS mRNA) prevents reuse of DsrA. Hfq is eventually recycled as a consequence of DsrA·rpoS base-pairing and/or by CsdA action. The segment of the iss is in red. Scissors signify RNase III cleavage. All other components are denoted.