Figures & data
Figure 1 NC's involvements in HIV-1 replicative cycle. (1) Gag proteins are first produced and migrate through the cytoplasm towards the vicinity of the cellular membrane. N-terminal MA domains of Gag bind to the lipid raft domains of the cellular membrane while the C-termini project into the cytoplasm and bind viral RNA through their NC domains. Gag binding initiates at the Ψ RNA region ensuring viral RNA dimerization, which then acts as a nucleation point to aggregate other Gag and Gag-Pol units though Gag-Gag interactions and NC/RNA binding (MA/RNA binding has also been proposedCitation62–Citation64). Viral RNA capture competes with the translational and/or mRNA docking apparati whereas other viral components are loaded onto Gag such as Vpr. (2) RNA scaffolding and Gag-Gag interactions mediate assembly of the particle which subsequently buds from the cell as an immature and imperfect particle. (3) Contacts between Gag-Pol precursors drive PR dimerization/autoactivation, which initiates virus maturation. (4) Sequential proteolytic events of Gag and Gag-Pol by PR and the self-assembly capabilities of the cleavage products (MA, CA, NC) reorganize the viral core, generating a mature particle. Maturation results in NC/RNA complex (i.e., the nucleocapsid) condensation at the center of the core where NC chaperone activity modulates RNA tertiary and quaternary structures. (5) After entry of the HIV-1 particle into the cytoplasm of a newly infected cell, RT catalyses viral DNA synthesis in the context of a ribonucleoproteic complex where NC proteins accelerate the strand transfers required for (−) and (+) strand synthesis. Viral dsDNA synthesis proceeds with a complete remodeling of the capsid and its contents, which are ultimately converted into a pre-integration complex, depleted of most of the NC initially bound to the viral RNA.
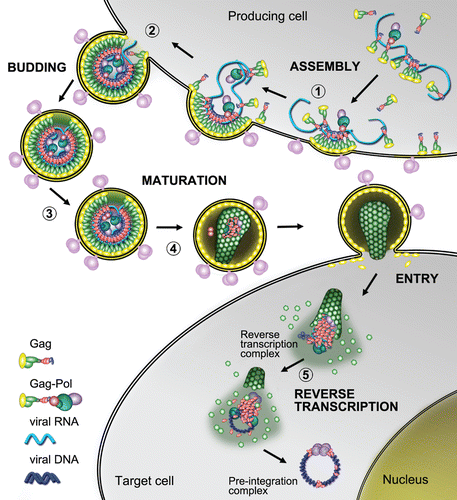
Figure 2 Structural features of HIV-1, RSV and MuLV nucleocapsid proteins—Ψ RNA complexes. The figure focuses on the binding of key guanosine residues (red), by a motif formed by a pocket derived from the side chains of hydrophobic residues (green). Zinc atoms bound by the ZFs are grey spheres. Location of basic amino-acids (Lys and Arg), available for electrostatic binding with RNA, are highlighted in yellow. (A) HIV-1 NC complexed with SL3 Ψ RNA (PDB ID:1A1T). A guanosine residue fits between the two adjacent ZFs which form a hydrophobic pocket around Phe16 and Trp37. The N-terminal basic tail adopts a 310 helix, which binds to the major groove of the SL3 stem.Citation16 (B) RSV NC and µΨ RNA structure (PDB ID:2IHX), where the guanosine base fits into a hydrophobic pocket defined by residues Tyr22, Tyr30, Leu20 and Gln31. Note that the RSV protein contains two ZF domains that adopt canonical folds observed for all other structurally characterized retroviral ZF, Tyr residues are used by RSV ZF1 instead of Trp and Phe residues used by HIV-1 NC. The C-terminal ZF is unusual as it does not contain aromatic residues nor a well-defined hydrophobic pocket. This finger packs against an adenine nucleobase though hydrogen bonds and salt bridges.Citation123 (C) Complex of MuLV NC and the mΨ RNA core encapsidation signal (PDB ID:1U6P) in which the single CCHC ZF of NC interacts with the UCUG sequence (indicated in pink, with G in red). The guanosine base fits deeply into a pocket defined by the side chain of Leu21, Ala27, Trp35 and Ala36. The first U and C pack against the side chain of Tyr28 and the second U packs against the side chains of Ala27 and Leu 21. The binding also appears to be promoted by several interactions between the basic residues and the phosphodiester backbone.Citation56
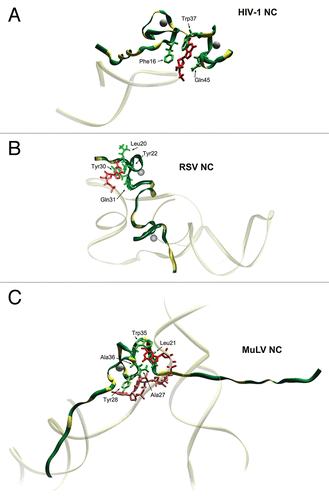
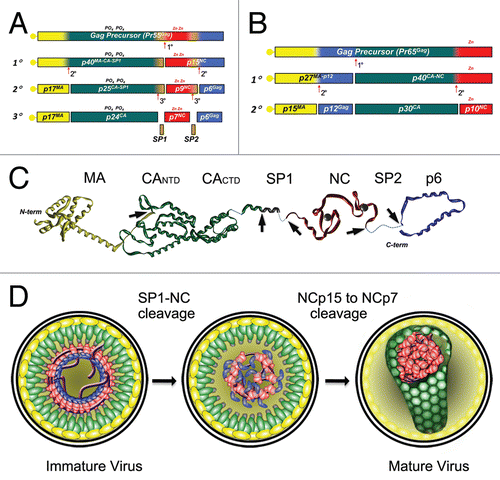
Figure 3 Proteolytic processing of HIV-1 and MuLV Gag by PR. The different protein species that exist during the steps of processing are indicated. (A) In HIV-1 Gag the initial cleavage occurs between SP1 and the NC domain, secondary cleavages occur at approximately 10-fold lower rates than the initial cut. The tertiary cleavages occur approximately 400-fold slower than the primary cleavage.Citation72 During proteolysis, HIV-1 NC exists in two intermediate forms, NCp15 (partial cleavage product containing NC/SP2/p6) and NCp9 (partial cleavage product containing NC/SP2) and the fully processed form, NCp7. All three of these proteins exhibit nucleic acid chaperone activities.Citation17 (B) Primary cleavage of MuLV Gag occurs between p12 and CA with slower secondary cleavages that yield the mature proteins MAp15, p12, CAp30 and NCp10.Citation124 (C) Extended structural model of an HIV-1 Gag polypeptide assembled from high resolution structures of isolated domains (PDB IDs:1UPH, 3GV2, 1A1T, 2C55 respectively for MA, CA, NCp7 and p6). Unavailable and/or unstructured domains are represented by dashed lines. PR cleavage site are indicated by the arrowheads. (D) Schematic models of viral core maturation. SP1-NC cleavage by PR rapidly separates the MA-CA shell and the nucleocapsid complex formed between RNA, NCp15, RT and IN. NCp15 processing by PR into NCp9 and finally NCp7 leads to NC/RNA coaggregation/condensation within the confines of the capsid cone.