Figures & data
Figure 1 Schematic illustration of the nucleic acids binding, fraying and annealing properties of the HIV nucleocapsid protein. Top, from left to right: NC protein is represented by an oval corresponding to the hydrophobic plateau (ZnF), flanked by the disordered N- and C-terminal domains shown here as small fragments. The 3D structure of the hydrophobic plateau at the top of the NC zinc-fingers is represented in the CPK mode with Val13, F16, Ile24, Ala25, Trp37, Gln45 and Met46, and the zinc atoms appear as grey spheres. The TAR with its loop and bulges is shown here as a stem-loop. The complementary DNA sequence is synthesized by RT chaperoned by NC at the very beginning of reverse transcription (reviewed in ref. Citation13 and Citation14) and shown here as cTAR stem-loop. Bottom, from left to right: Binding of NC to TAR results in the formation of a ribonucleoparticle where the RNA is entirely coated by NC molecules. This causes the transient opening of the 5′ and 3′ end sequences (short arrows), in a process called fraying and strand stretching (Single DNA molecule stretching measures the activity of chemicals that target the HIV-1 nucleocapsid protein).Citation53 Binding of NC to TAR and cTAR causes the formation of nucleoprotein complexes where the nucleic acids are entirely coated by NC molecules. This causes the rapid hybridization of the complementary sequences resulting in the formation of a RNA:DNA hybrid within the nucleoprotein complex. This annealing reaction recapitulates the obligatory 5′-3′ strand transfer that occur at the beginning of reverse transcription.Citation4
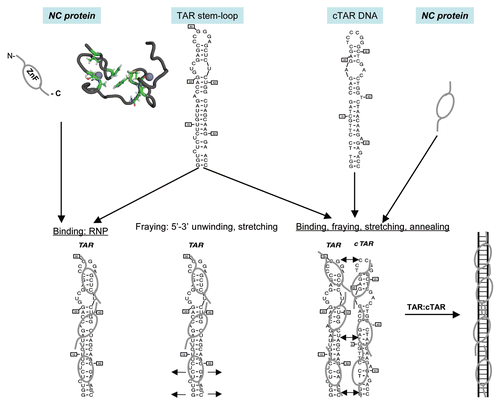
Figure 2 The late steps of retroviral particle formation: traffic, assembly and budding. The different stages of NC: immature NC in Gag and mature NC in the viral particle. Scheme of Gag-gRNA complexes trafficking within the cell and viral particle assembly and release. Upon export of the genomic RNA from the nucleus, the gRNA is translated into Gag and GagPol proteins that recruit the genome and traffic through the cytoplasm onto the assembly site et the plasma membrane; few Gag complexes with the viral genome anchor into the plasma membrane; Gag recognizes the PM phospholipids PI(4,5)P2 which triggers the anchoring of the myristate into the inner leaflet of the membrane, multimers of Gag thus accumulate at the assembly site which can be rafts or tetraspanins enriched microdomains and will serve as a plateform for particle budding; other cellular determinant can intervene to promote budding and particle release, such as the ESCRT machinery. Some preformed or formed virions can undergo endocytosis from the plasma membrane and create new virus-containing compartment (VVC) in the cell that could be released by exocytosis upon cellular signal. During release, the particle undergoes a maturation process due to the viral protease that cuts Gag and GagPol to specific sites which releases the structural proteins and enzymes in order to form the mature infectious particle, in which free NC proteins condensed on the genomic RNA inside the viral mature core. This phenomenon is a key determinant for particle morphogenesis and infectivity.
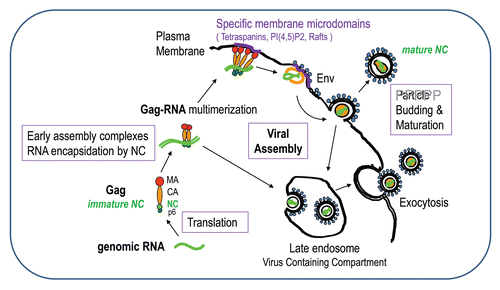
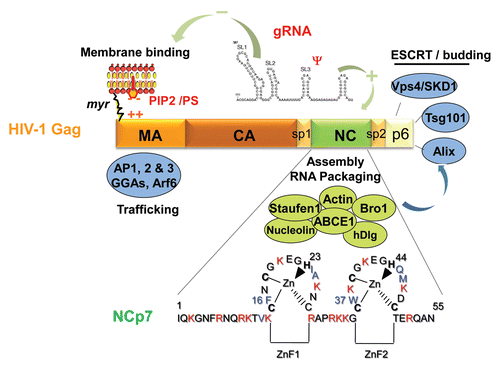
Figure 3 The Gag polyprotein precursor, its NC domain and the associated cellular factors. HIV-1 Gag is composed of 3 main domains which are MA, CA and NC, one C-terminal peptide p6 and two spacer peptides, sp1 and sp2. The matrix MA domain is responsible for Gag membrane binding via the myristate and the basic (++) region with the PIP2/PS phospholipids. MA domain contains motifs that bind different cellular factors, such as AP1, AP2, AP3, GGA and Arf6 involved in Gag trafficking. The NC domain of Gag is necessary for gRNA recruitment via its interaction with the Psi packaging signal and the NC basic residues (in red) and the hydrophobic plateau encompassing the two conserved zinc fingers. The gRNA is also able to interact with the MA domain by competing with PI(4,5)P2. The NC domain is a key factor for the early assembly complex formation by Gag multimerization on the gRNA. NC and p6 interact with cellular factors; NC with actin, Staufen1, the Bro1 domain of ALIX and ABCE1 as described in this review; the C-terminus of Gag, via the p6 peptide containing the late budding domain is able to recruit the ESCRT cellular proteins, such as Tsg101, Alix and Vps4/SDK1, which are essential for particle budding. Sp1 and Sp2 are linker involved in HIV-1 morphogenesis. The 55 amino acids of mature NC are given; NCp7 is highly basic (in red) and contains two highly conserved zinc CCHC binding motifs and a plateau of hydrophobic amino-acids (in blue) involved in RNA-NC interactions, RNA chaperone activities and particle assembly.