Figures & data
Figure 1 Heat map of the 33 differentially expressed miRNAs between the CEU and YRI samples. Each column represents a sample with CEU samples placed on the left half of the heat map (n = 53); and YRI samples on the right half of the heat map (n = 54). Each row contains normalized expression intensity of a differentially expressed miRNA (bonferroni corrected p < 0.05 with FDR ≤ 0.0005).
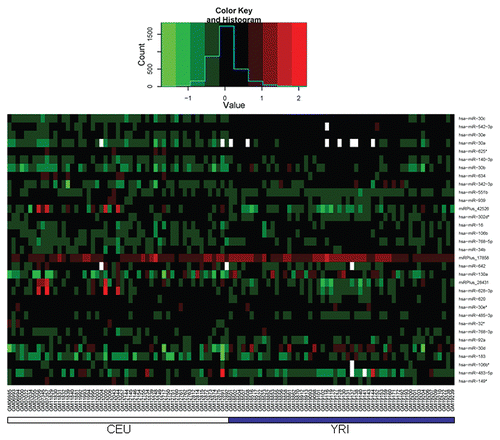
Figure 2 The expression of miR-30b, and ZNRF1 in CEU and YRI samples. (A and B) The expression of miR-30b and ZNRF1 in the discovery CEU I/II (open square) and YRI I/II (solid triangle) samples. miR-30b expression values were obtained via Exiqon array. ZNRF1 expression values were assessed using Affymetrix Exon array. (C and D) The expression of miR-30b, and ZNRF1 in the validation CEU III (open square) and YRI III (solid triangle) samples. miR-30b and ZNRF1 expression values were obtained via real-time PCR.
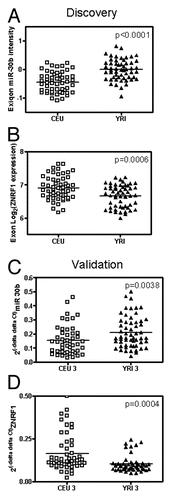
Figure 3 The relationship between the expression of miR-342-3p and ANKRD49 in CEU and YRI samples. (A) The relationship between the expression of miR-342-3p and ANKRD49 in the discovery CEU I/II (open square) and YRI I/II (solid triangle) samples. miR-342-3p expression values were obtained via Exiqon array. ANKRD49 expression values were assessed using Affymetrix Exon array. (B) The relationship between expression of miR-342-3p and ANKRD49 in the validation CEU III and YRI III samples. miR-342-3p and ANKRD49 expression values were obtained via real-time PCR.
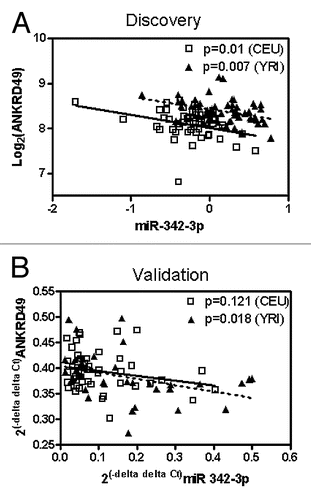
Figure 4 Effect of miRNA mimics and inhibitors on the expression of miRNAs and target genes. The x axis describes the type of experiment. The y axis illustrates the percent change in expression level when compared with scramble control in each experiment either for miRNAs or for target genes.
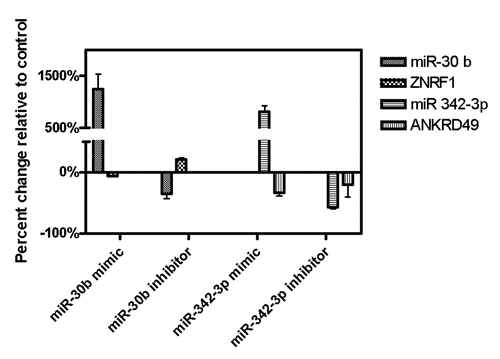
Table 1 The inverse correlation between differentially expressed miRNAs and mRNAs in CEU and YRI samples
Table 2 The relationship between differentially expressed miRNAs and chemo-sensitivity in CEU and YRI samples