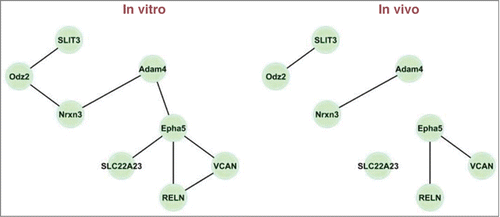
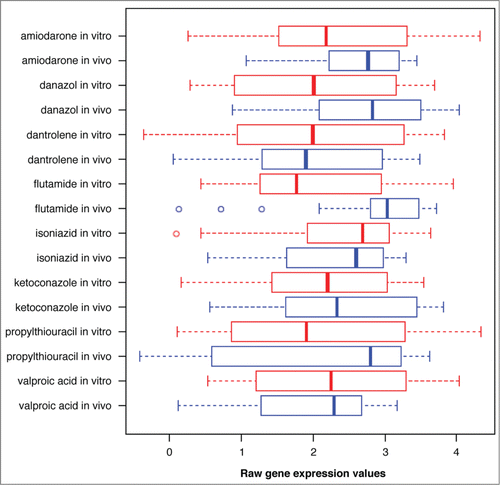
Figures & data
Table 1. Drugs with similar connectivity scores in the two networks are provided
Table 2. Genes which are not differentially expressed between the networks for at least 80% of the 18 high DILI drugs for which there is a statistically significant difference between the overall connectivity score for in vivo vs. in vitro networks based on the set of all genes
Table 3. The top 5 clusters obtained from the DAVID Functional Annotation Tool
Table 4A. Proportions of high DILI drugs for which the downregulated genes from Guzelian et al. common to the TGP data set are not differentially expressed
Table 4B. Proportions of high DILI drugs for which the differentially expressed genes (between cirrhotic and control groups) from Tugues et al. common to the TGP data set are not differentially expressed