Figures & data
Figure 1. Cloverleaf folding of tRNA (tRNA). The standard cloverleaf structure (2D) and conventional numbering is shown. Conserved nucleotides are indicated (T and Ψ are modified residues: ribothymidine and pseudouridine; R are for purines and Y for pyrimidines). Watson-Crick base pairings and G•U pairs are shown; thin black lines indicate long-distance base pairings involved in tertiary folding (3D, upper right panel).
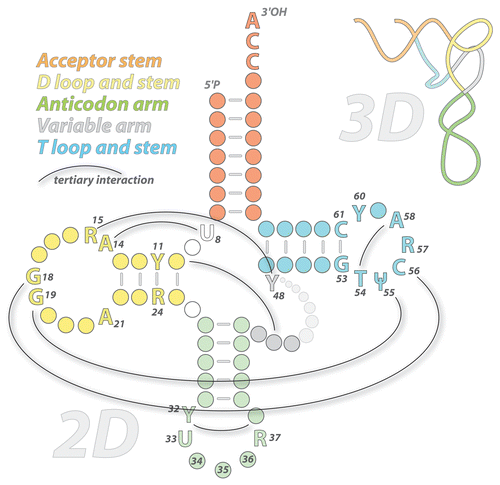
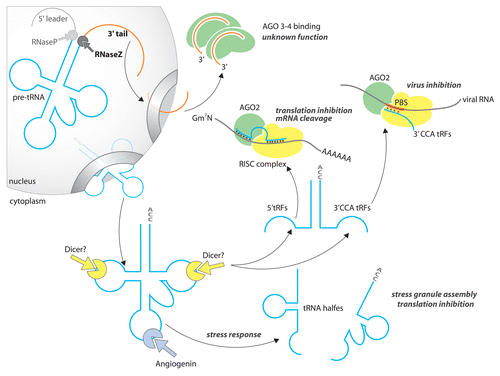
Figure 2. Synthesis, processing, and nucleo-cytoplasmic export of eukaryotic tRNA. In eukaryotes, tRNA concentrations fluctuate according to cell demand and are not proportional to the number tRNA genes in the genome (1). tRNA genes are transcribed by RNA polymerase (2) as long precursors (pre-tRNA). Additional sequences are then enzymatically removed (3). Functional tRNAs display a CCA sequence at their 3′-end. This triplet is added post-transcriptionally by the tRNA nucleotidyl transferase (CCase) (4). Base modifications are added at the final processing step (5). Export of tRNA through nuclear pores is facilitated by exportin-t and Ran-GTP (6).
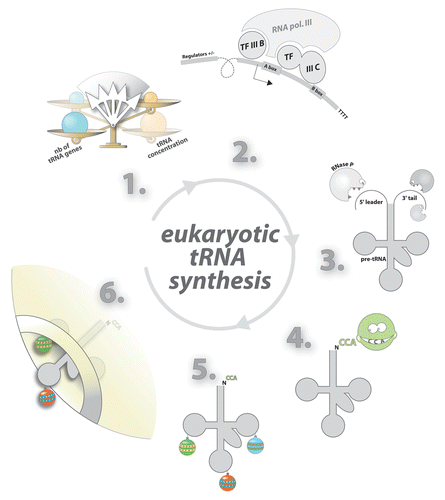
Figure 3. Number of tRNA genes and isodecoder genes across the phylogenetic spectrum. Eukaryotic genomes contain between 200 and 450 tRNA genes encoding 41 to 55 tRNA isoacceptors (tRNAs with different anticodons). The number of tRNA genes having the same anticodon but different sequences elsewhere in the tRNA body (tRNA isodecoders) varies significantly and is increasing across the phylogenetic spectrum. Sequence variations in tRNA isodecoders are concentrated in the internal promoter regions for RNA polymerase III.
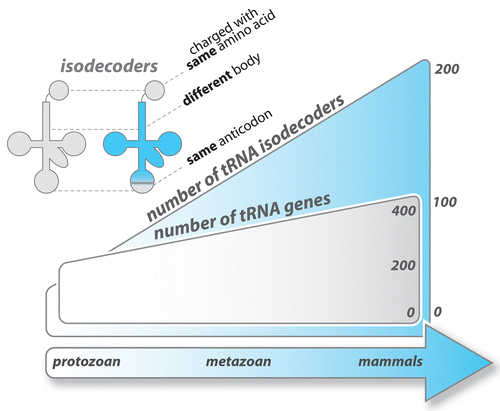
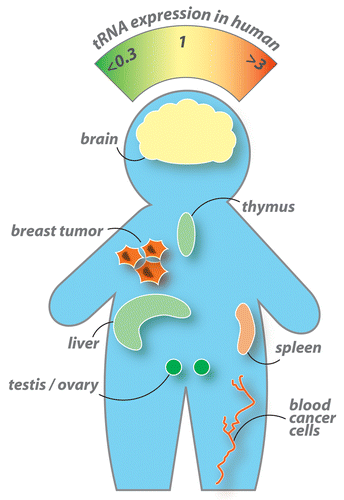
Figure 4. Differential tRNA expression in seven different human tissues measured by tRNA microarrays. Tumor cells generally express higher levels of nuclear encoded tRNA than every tissue examined.