Figures & data
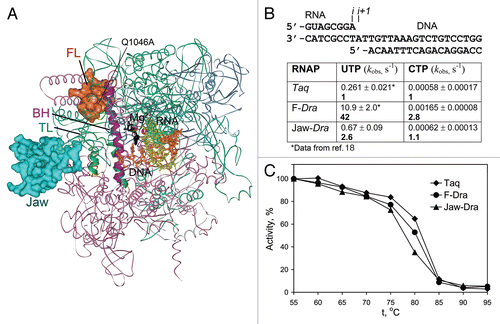
Figure 1 Structure of the RNAP active center and conformational transitions of the TL. (A) and (B) Structures of the Tth transcription elongation complex (TEC) with NTP (black) in the i+1 siteCitation2 and Tth holoenzyme RNAP (a core bound to the promoter-specific σ subunit),Citation28 respectively. Catalytic Mg2+ ions are shown as red spheres, the template DNA is in orange, RNA is in yellow, the FL is in red, the BH is in magenta, the TL is in green. Q1046, S1074 and H1242 in the FL, BH and TL are shown as CPK models in yellow, light violet and dark green, respectively. Light green spheres indicates position of the G1136SEco substitution; blue spheres indicate positions of the M747IEco, P750LEco, F773VEco and R780HEco substitutions; gray spheres—insertions in the TL in Eco RNAP (ΩSI3) and the FL (ΩFL) in various bacterial lineages. A part of the jaw-domain (semitransparent in A) is not visible in the TEC structure; variable Taq/Dra residues at the base of this domain are in turquoise. (C) and (D) Structures of the Sce TEC with NTP in the i+1 siteCitation1 and RNAPII bound to TFIIS.Citation25 The TL E1103Sce and an adjacent K1112Sce in the jaw-domain are in light green and turquoise, respectively; S769Sce (yellow) in the FL corresponds to Q1046Taq. Positions of sit-substitutions (blue) and hyperactive Mja variants in the BH (pink) and the TL (light green) are indicated. The Zn-ribbon domains of Rpb9 and TFIIS are dark blue and gray, respectively; Rpb9 K93 residue (a dark blue sphere) is adjacent to the open TL. (E) Structure of a backtracked Sce TECCitation11 (RNA 3′-nucleotide is in black, the TL is partially open) and of TEC in complex with α-amanitin (black)Citation3 with the TL in a wedged conformation.
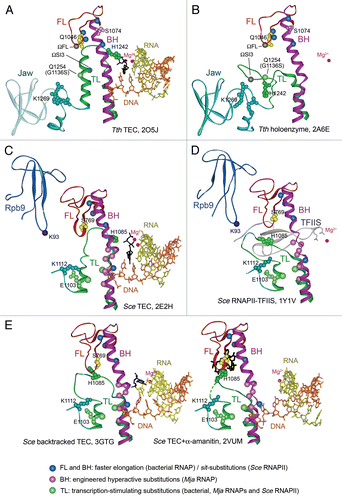
Figure 2 Transcription properties of the mosaic RNAP variants. (A) Positions of the β′ substitutions in mosaic RNAPs shown on the structure of Tth TEC in complex with Stl (2PPBCitation2). RNAP orientation corresponds to . The β (pink), β′ (green), α (blue) and ω (gray) subunits are shown as ribbons; the FL and the jaw-domain are in red and turquoise, respectively. (B) Rates of single nucleotide addition by mosaic Taq TECs assembled on the minimal scaffold (top) were measured at 20°C and 1 mM NTP as described previously.Citation18 Numbers in bold indicate fold differences in comparison with wild-type Taq RNAP. (C) Thermostability of mosaic Taq RNAPs. RNAPs were incubated for 3 min at indicated temperatures and their activity was measured on the T7A1 promoter in the presence of a CpA primer and UTP (10 µM each) for 5 min at 55°C.